Performance and Prediction¶
Model Performance¶
Given a trained H2O model, the h2o.performance() (R)/model_performance() (Python) function computes a model’s performance on a given dataset.
Notes:
- If the provided dataset does not contain the response/target column from the model object, no performance will be returned. Instead, a warning message will be printed.
- For binary classification problems, H2O uses the model along with the given dataset to calculate the threshold that will give the maximum F1 for the given dataset.
This section describes how H2O-3 can be used to evaluate model performance. Models can also be evaluated with specific model metrics, stopping metrics, and performance graphs.
Evaluation Model Metrics¶
H2O-3 provides a variety of metrics that can be used for evaluating supervised and unsupervised models. The metrics for this section only cover supervised learning models, which vary based on the model type (classification or regression).
Regression¶
The following evaluation metrics are available for regression models. (Note that H2O-3 also calculates regression metrics for Classification problems.)
- R2 (R Squared)
- MSE (Mean Squared Error)
- RMSE (Root Mean Squared Error)
- RMSLE (Root Mean Squared Logarithmic Error)
- MAE (Mean Absolute Error)
Each metric is described in greater detail in the sections that follow. The examples are based off of a GBM model built using the cars_20mpg.csv dataset.
library(h2o)
h2o.init()
# import the cars dataset:
cars <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the predictor names and the response column name
predictors <- c("displacement","power","weight","acceleration","year")
response <- "cylinders"
# split into train and validation sets
cars.splits <- h2o.splitFrame(data = cars, ratios = .8, seed = 1234)
train <- cars.splits[[1]]
valid <- cars.splits[[2]]
# build and train the model:
cars.gbm <- h2o.gbm(x = predictors,
y = response,
training_frame = train,
validation_frame = valid,
distribution = "poisson",
seed = 1234)
# retrieve the model performance
perf <- h2o.performance(cars.gbm, valid)
perf
import h2o
from h2o.estimators.gbm import H2OGradientBoostingEstimator
h2o.init()
# import the cars dataset:
# this dataset is used to classify whether or not a car is economical based on
# the car's displacement, power, weight, and acceleration, and the year it was made
cars = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the predictor names and the response column name
predictors = ["displacement","power","weight","acceleration","year"]
response = "cylinders"
# split into train and validation sets
train, valid = cars.split_frame(ratios = [.8], seed = 1234)
# train a GBM model
cars_gbm = H2OGradientBoostingEstimator(distribution = "poisson", seed = 1234)
cars_gbm.train(x = predictors,
y = response,
training_frame = train,
validation_frame = valid)
# retrieve the model performance
perf = cars_gbm.model_performance(valid)
perf
R2 (R Squared)¶
The R2 value represents the degree that the predicted value and the actual value move in unison. The R2 value varies between 0 and 1 where 0 represents no correlation between the predicted and actual value and 1 represents complete correlation.
Example
Using the previous example, run the following to retrieve the R2 value.
# retrieve the r2 value:
r2.basic <- h2o.r2(cars.gbm)
r2.basic
[1] 0.9930651
# retrieve the r2 value for the validation data:
r2.basic_valid <- h2o.r2(cars.gbm, valid=TRUE)
r2.basic_valid
[1] 0.9886704
# retrieve the r2 value:
cars_gbm.r2()
0.9930650688408735
# retrieve the r2 value for the validation data:
cars_gbm.r2(valid=True)
0.9886704207301097
MSE (Mean Squared Error)¶
The MSE metric measures the average of the squares of the errors or deviations. MSE takes the distances from the points to the regression line (these distances are the “errors”) and squaring them to remove any negative signs. MSE incorporates both the variance and the bias of the predictor.
MSE also gives more weight to larger differences. The bigger the error, the more it is penalized. For example, if your correct answers are 2,3,4 and the algorithm guesses 1,4,3, then the absolute error on each one is exactly 1, so squared error is also 1, and the MSE is 1. But if the algorithm guesses 2,3,6, then the errors are 0,0,2, the squared errors are 0,0,4, and the MSE is a higher 1.333. The smaller the MSE, the better the model’s performance. (Tip: MSE is sensitive to outliers. If you want a more robust metric, try mean absolute error (MAE).)
MSE equation:
\[MSE = \frac{1}{N} \sum_{i=1}^{N}(y_i -\hat{y}_i)^2\]
Example
Using the previous example, run the following to retrieve the MSE value.
# retrieve the mse value:
mse.basic <- h2o.mse(cars.gbm)
mse.basic
[1] 0.01917327
# retrieve the mse value for both the training and validation data:
mse.basic_valid <- h2o.mse(cars.gbm, train=TRUE, valid=TRUE, xval=FALSE)
mse.basic_valid
train valid
0.01917327 0.03769792
# retrieve the mse value:
cars_gbm.mse()
0.019173269728097173
# retrieve the mse value for the validation data:
cars_gbm.mse(valid=True)
0.03769791966551617
RMSE (Root Mean Squared Error)¶
The RMSE metric evaluates how well a model can predict a continuous value. The RMSE units are the same as the predicted target, which is useful for understanding if the size of the error is of concern or not. The smaller the RMSE, the better the model’s performance. (Tip: RMSE is sensitive to outliers. If you want a more robust metric, try mean absolute error (MAE).)
RMSE equation:
\[RMSE = \sqrt{\frac{1}{N} \sum_{i=1}^{N}(y_i -\hat{y}_i)^2 }\]
Where:
- N is the total number of rows (observations) of your corresponding dataframe.
- y is the actual target value.
- \(\hat{y}\) is the predicted target value.
Example
Using the previous example, run the following to retrieve the RMSE value.
# retrieve the rmse value:
rmse.basic <- h2o.rmse(cars.gbm)
rmse.basic
[1] 0.1384676
# retrieve the rmse value for both the training and validation data:
rmse.basic_valid <- h2o.rmse(cars.gbm, train=TRUE, valid=TRUE, xval=FALSE)
rmse.basic_valid
train valid
0.1384676 0.1941595
# retrieve the rmse value:
cars_gbm.rmse()
0.13846757645057983
# retrieve the rmse value for the validation data:
cars_gbm.rmse(valid=True)
0.19415952118172358
RMSLE (Root Mean Squared Logarithmic Error)¶
This metric measures the ratio between actual values and predicted values and takes the log of the predictions and actual values. Use this instead of RMSE if an under-prediction is worse than an over-prediction. You can also use this when you don’t want to penalize large differences when both of the values are large numbers.
RMSLE equation:
\[RMSLE = \sqrt{\frac{1}{N} \sum_{i=1}^{N} \big(ln \big(\frac{y_i +1} {\hat{y}_i +1}\big)\big)^2 }\]
Where:
- N is the total number of rows (observations) of your corresponding dataframe.
- y is the actual target value.
- \(\hat{y}\) is the predicted target value.
Example
Using the previous example, run the following to retrieve the RMSLE value.
# retrieve the rmsle value:
rmsle.basic <- h2o.rmsle(cars.gbm)
rmsle.basic
[1] 0.02332083
# retrieve the rmsle value for both the training and validation data:
rmsle.basic_valid <- h2o.rmsle(cars.gbm, train=TRUE, valid=TRUE, xval=FALSE)
rmsle.basic_valid
train valid
0.02332083 0.03359130
# retrieve the rmsle value:
cars_gbm.rmsle()
0.023320830800314333
# retrieve the rmsle value for the validation data:
cars_gbm.rmsle(valid=True)
0.03359130162278705
MAE (Mean Absolute Error)¶
The mean absolute error is an average of the absolute errors. The MAE units are the same as the predicted target, which is useful for understanding whether the size of the error is of concern or not. The smaller the MAE the better the model’s performance. (Tip: MAE is robust to outliers. If you want a metric that is sensitive to outliers, try root mean squared error (RMSE).)
MAE equation:
\[MAE = \frac{1}{N} \sum_{i=1}^{N} | x_i - x |\]
Where:
- N is the total number of errors
- \(| x_i - x |\) equals the absolute errors.
Example
Using the previous example, run the following to retrieve the MAE value.
# retrieve the mae value:
mae.basic <- h2o.mae(cars.gbm)
mae.basic
[1] 0.06140515
# retrieve the mae value for both the training and validation data:
mae.basic_valid <- h2o.mae(cars.gbm, train=TRUE, valid=TRUE, xval=FALSE)
mae.basic_valid
train valid
0.06140515 0.07947862
# retrieve the mae value:
cars_gbm.mae()
0.06140515094616347
# retrieve the mae value for the validation data:
cars_gbm.mae(valid=True)
0.07947861719967757
Classification¶
H2O-3 calculates regression metrics for classification problems. The following additional evaluation metrics are available for classification models:
- Gini Coefficient
- Absolute MCC (Matthews Correlation Coefficient)
- F1
- F0.5
- F2
- Accuracy
- Logloss
- AUC (Area Under the ROC Curve)
- AUCPR (Area Under the Precision-Recall Curve)
Each metric is described in greater detail in the sections that follow. The examples are based off of a GBM model built using the allyears2k_headers.zip dataset.
library(h2o)
h2o.init()
# import the airlines dataset:
# This dataset is used to classify whether a flight will be delayed 'YES' or not "NO"
# original data can be found at http://www.transtats.bts.gov/
airlines <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
# convert columns to factors
airlines["Year"] <- as.factor(airlines["Year"])
airlines["Month"] <- as.factor(airlines["Month"])
airlines["DayOfWeek"] <- as.factor(airlines["DayOfWeek"])
airlines["Cancelled"] <- as.factor(airlines["Cancelled"])
airlines['FlightNum'] <- as.factor(airlines['FlightNum'])
# set the predictor names and the response column name
predictors <- c("Origin", "Dest", "Year", "UniqueCarrier",
"DayOfWeek", "Month", "Distance", "FlightNum")
response <- "IsDepDelayed"
# split into train and validation
airlines.splits <- h2o.splitFrame(data = airlines, ratios = .8, seed = 1234)
train <- airlines.splits[[1]]
valid <- airlines.splits[[2]]
# build a model
airlines.gbm <- h2o.gbm(x = predictors,
y = response,
training_frame = train,
validation_frame = valid,
sample_rate =.7,
seed = 1234)
# retrieve the model performance
perf <- h2o.performance(airlines.gbm, valid)
perf
import h2o
from h2o.estimators.gbm import H2OGradientBoostingEstimator
h2o.init()
# import the airlines dataset:
# This dataset is used to classify whether a flight will be delayed 'YES' or not "NO"
# original data can be found at http://www.transtats.bts.gov/
airlines= h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
# convert columns to factors
airlines["Year"]= airlines["Year"].asfactor()
airlines["Month"]= airlines["Month"].asfactor()
airlines["DayOfWeek"] = airlines["DayOfWeek"].asfactor()
airlines["Cancelled"] = airlines["Cancelled"].asfactor()
airlines['FlightNum'] = airlines['FlightNum'].asfactor()
# set the predictor names and the response column name
predictors = ["Origin", "Dest", "Year", "UniqueCarrier",
"DayOfWeek", "Month", "Distance", "FlightNum"]
response = "IsDepDelayed"
# split into train and validation sets
train, valid = airlines.split_frame(ratios = [.8], seed = 1234)
# train your model
airlines_gbm = H2OGradientBoostingEstimator(sample_rate = .7, seed = 1234)
airlines_gbm.train(x = predictors,
y = response,
training_frame = train,
validation_frame = valid)
# retrieve the model performance
perf = airlines_gbm.model_performance(valid)
perf
Gini Coefficient¶
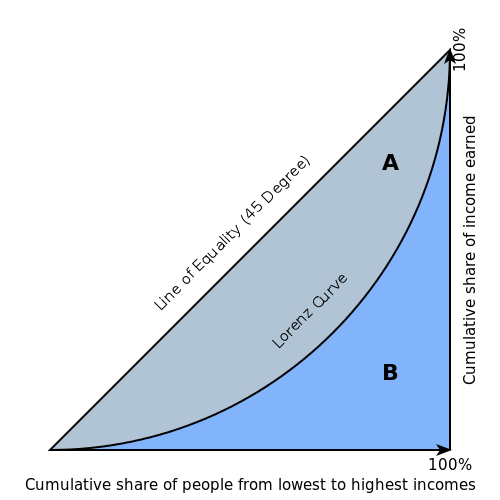
The Gini index is a well-established method to quantify the inequality among values of a frequency distribution, and can be used to measure the quality of a binary classifier. A Gini index of zero expresses perfect equality (or a totally useless classifier), while a Gini index of one expresses maximal inequality (or a perfect classifier).
The Gini index is based on the Lorenz curve. The Lorenz curve plots the true positive rate (y-axis) as a function of percentiles of the population (x-axis).
The Lorenz curve represents a collective of models represented by the classifier. The location on the curve is given by the probability threshold of a particular model. (i.e., Lower probability thresholds for classification typically lead to more true positives, but also to more false positives.)
The Gini index itself is independent of the model and only depends on the Lorenz curve determined by the distribution of the scores (or probabilities) obtained from the classifier.
Example
Using the previous example, run the following to retrieve the Gini coefficient value.
# retrieve the gini value for the performance object:
h2o.giniCoef(perf)
[1] 0.482994
# retrieve the gini value for both the training and validation data:
h2o.giniCoef(airlines_gbm, train=TRUE, valid=TRUE, xval=FALSE)
train valid
0.5715841 0.4829940
# retrieve the gini coefficient:
perf.gini()
0.48299402265152613
# retrieve the gini coefficient for both the training and validation data:
airlines_gbm.gini(train=True, valid=True, xval=False)
{u'train': 0.5715841348613386, u'valid': 0.48299402265152613}
Absolute MCC (Matthews Correlation Coefficient)¶
Setting the absolute_mcc parameter sets the threshold for the model’s confusion matrix to a value that generates the highest Matthews Correlation Coefficient. The MCC score provides a measure of how well a binary classifier detects true and false positives, and true and false negatives. The MCC is called a correlation coefficient because it indicates how correlated the actual and predicted values are; 1 indicates a perfect classifier, -1 indicates a classifier that predicts the opposite class from the actual value, and 0 means the classifier does no better than random guessing.
Example
Using the previous example, run the following to retrieve the MCC value.
# retrieve the mcc value for the performance object:
h2o.mcc(perf)
threshold absolute_mcc
1 0.9636255 0.01754051
2 0.9590688 0.03509912
3 0.9536574 0.03924877
4 0.9510736 0.04862323
5 0.9488456 0.05738251
---
threshold absolute_mcc
395 0.10401437 0.04106864
396 0.09852580 0.03994376
397 0.09265314 0.03664277
398 0.08816490 0.02184613
399 0.06793601 0.01960485
400 0.06432841 0.00000000
# retrieve the mcc for the performance object:
perf.mcc()
[0.5426977730968023, 0.36574105494931725]]
# retrieve the mcc for both the training and validation data:
airlines_gbm.mcc(train=True, valid=True, xval=False)
{u'train': [[0.5203060957871319, 0.42414048381779923]], u'valid': [[0.5426977730968023, 0.36574105494931725]]}
F1¶
The F1 score provides a measure for how well a binary classifier can classify positive cases (given a threshold value). The F1 score is calculated from the harmonic mean of the precision and recall. An F1 score of 1 means both precision and recall are perfect and the model correctly identified all the positive cases and didn’t mark a negative case as a positive case. If either precision or recall are very low it will be reflected with a F1 score closer to 0.
Where:
- precision is the positive observations (true positives) the model correctly identified from all the observations it labeled as positive (the true positives + the false positives).
- recall is the positive observations (true positives) the model correctly identified from all the actual positive cases (the true positives + the false negatives).
Example
Using the previous example, run the following to retrieve the F1 value.
# retrieve the F1 value for the performance object:
h2o.F1(perf)
threshold f1
1 0.9636255 0.001301801
2 0.9590688 0.005197055
3 0.9536574 0.006492101
4 0.9510736 0.009937351
5 0.9488456 0.013799051
---
threshold f1
395 0.10401437 0.6916548
396 0.09852580 0.6915972
397 0.09265314 0.6914934
398 0.08816490 0.6911301
399 0.06793601 0.6910728
400 0.06432841 0.6909173
# retrieve the F1 coefficient for the performance object:
perf.F1()
[[0.35417599264806404, 0.7228980805623143]]
# retrieve the F1 coefficient for both the training and validation data:
airlines_gbm.F1(train=True, valid=True, xval=False)
{u'train': [[0.3869697386893616, 0.7451099672437997]], u'valid': [[0.35417599264806404, 0.7228980805623143]]}
F0.5¶
The F0.5 score is the weighted harmonic mean of the precision and recall (given a threshold value). Unlike the F1 score, which gives equal weight to precision and recall, the F0.5 score gives more weight to precision than to recall. More weight should be given to precision for cases where False Positives are considered worse than False Negatives. For example, if your use case is to predict which products you will run out of, you may consider False Positives worse than False Negatives. In this case, you want your predictions to be very precise and only capture the products that will definitely run out. If you predict a product will need to be restocked when it actually doesn’t, you incur cost by having purchased more inventory than you actually need.
F0.5 equation:
\[F0.5 = 1.25 \;\Big(\; \frac{(precision) \; (recall)}{0.25 \; precision + recall}\; \Big)\]
Where:
- precision is the positive observations (true positives) the model correctly identified from all the observations it labeled as positive (the true positives + the false positives).
- recall is the positive observations (true positives) the model correctly identified from all the actual positive cases (the true positives + the false negatives).
Example
Using the previous example, run the following to retrieve the F0.5 value.
# retrieve the F0.5 value for the performance object:
h2o.F0point5(perf)
threshold f0point5
1 0.9636255 0.003248159
2 0.9590688 0.012892136
3 0.9536574 0.016073725
4 0.9510736 0.024478501
5 0.9488456 0.033798057
---
threshold f0point5
395 0.10401437 0.5837602
396 0.09852580 0.5836502
397 0.09265314 0.5835319
398 0.08816490 0.5831181
399 0.06793601 0.5830085
400 0.06432841 0.5828314
# retrieve the F1 coefficient for the performance object:
perf.F0point5()
[[0.5426977730968023, 0.7047449127206096]]
# retrieve the F1 coefficient for both the training and validation data:
airlines_gbm.F0point5(train=True, valid=True, xval=False)
{u'train': [[0.5529885092975969, 0.7331482319556736]], u'valid': [[0.5426977730968023, 0.7047449127206096]]}
F2¶
The F2 score is the weighted harmonic mean of the precision and recall (given a threshold value). Unlike the F1 score, which gives equal weight to precision and recall, the F2 score gives more weight to recall (penalizing the model more for false negatives then false positives). An F2 score ranges from 0 to 1, with 1 being a perfect model.
Example
Using the previous example, run the following to retrieve the F2 value.
# retrieve the F2 value for the performance object:
h2o.F2(perf)
threshold f2
1 0.9636255 0.0008140229
2 0.9590688 0.0032545021
3 0.9536574 0.0040674657
4 0.9510736 0.0062340760
5 0.9488456 0.0086692674
---
threshold f2
395 0.10401437 0.8484759
396 0.09852580 0.8485351
397 0.09265314 0.8484726
398 0.08816490 0.8482538
399 0.06793601 0.8483130
400 0.06432841 0.8482192
# retrieve the F2 coefficient for the performance object:
perf.F2()
[[0.1957813426628461, 0.8502311018339048]]
# retrieve the F2 coefficient for both the training and validation data:
airlines_gbm.F2(train=True, valid=True, xval=False)
{u'train': [[0.24968434313831914, 0.8548787509793371]], u'valid': [[0.1957813426628461, 0.8502311018339048]]}
Accuracy¶
In binary classification, Accuracy is the number of correct predictions made as a ratio of all predictions made. In multiclass classification, the set of labels predicted for a sample must exactly match the corresponding set of labels in y_true.
Accuracy equation:
\[Accuracy = \Big(\; \frac{\text{number correctly predicted}}{\text{number of observations}}\; \Big)\]
Example
Using the previous example, run the following to retrieve the Accurace value.
# retrieve the Accuracy value for the performance object:
h2o.accuracy(perf)
threshold accuracy
1 0.9636255 0.4725564
2 0.9590688 0.4735877
3 0.9536574 0.4739315
4 0.9510736 0.4748482
5 0.9488456 0.4758795
---
threshold accuracy
395 0.10401437 0.5296207
396 0.09852580 0.5293915
397 0.09265314 0.5291624
398 0.08816490 0.5283603
399 0.06793601 0.5281311
400 0.06432841 0.5277873
# retrieve the accuracy coefficient for the performance object:
perf.accuracy()
[[0.5231232172827827, 0.6816775524235132]]
# retrieve the accuracy coefficient for both the training and validation data:
airlines_gbm.accuracy(train=True, valid=True, xval=False)
{u'train': [[0.5164521833040745, 0.7118095940540694]], u'valid': [[0.5231232172827827, 0.6816775524235132]]}
Logloss¶
The logarithmic loss metric can be used to evaluate the performance of a binomial or multinomial classifier. Unlike AUC which looks at how well a model can classify a binary target, logloss evaluates how close a model’s predicted values (uncalibrated probability estimates) are to the actual target value. For example, does a model tend to assign a high predicted value like .80 for the positive class, or does it show a poor ability to recognize the positive class and assign a lower predicted value like .50? Logloss can be any value greater than or equal to 0, with 0 meaning that the model correctly assigns a probability of 0% or 100%.
Binary classification equation:
\[Logloss = - \;\frac{1}{N} \sum_{i=1}^{N}w_i(\;y_i \ln(p_i)+(1-y_i)\ln(1-p_i)\;)\]
Multiclass classification equation:
\[Logloss = - \;\frac{1}{N} \sum_{i=1}^{N}\sum_{j=1}^{C}w_i(\;y_i,_j \; \ln(p_i,_j)\;)\]
Where:
- N is the total number of rows (observations) of your corresponding dataframe.
- w is the per row user-defined weight (defaults is 1).
- C is the total number of classes (C=2 for binary classification).
- p is the predicted value (uncalibrated probability) assigned to a given row (observation).
- y is the actual target value.
Example
Using the previous example, run the following to retrieve the logloss value.
# retrieve the logloss value for the performance object:
h2o.logloss(perf)
[1] 0.5967029
# retrieve the logloss value for both the training and validation data:
h2o.logloss(airlines.gbm, train=TRUE, valid=TRUE, xval=FALSE)
train valid
0.5607155 0.5967029
# retrieve the logloss for the performance object:
perf.gini()
0.5967028742962095
# retrieve the logloss for both the training and validation data:
airlines_gbm.logloss(train=True, valid=True, xval=False)
{u'train': 0.5607154587919981, u'valid': 0.5967028742962095}
AUC (Area Under the ROC Curve)¶
This model metric is used to evaluate how well a binary classification model is able to distinguish between true positives and false positives. An AUC of 1 indicates a perfect classifier, while an AUC of .5 indicates a poor classifier, whose performance is no better than random guessing. H2O uses the trapezoidal rule to approximate the area under the ROC curve.
H2O uses the trapezoidal rule to approximate the area under the ROC curve. (Tip: AUC is usually not the best metric for an imbalanced binary target because a high number of True Negatives can cause the AUC to look inflated. For an imbalanced binary target, we recommend AUCPR or MCC.)
Example
Using the previous example, run the following to retrieve the AUC.
# retrieve the AUC for the performance object:
h2o.auc(perf)
[1] 0.741497
# retrieve the AUC for both the training and validation data:
h2o.auc(airlines.gbm, train=TRUE, valid=TRUE, xval=FALSE)
train valid
0.7857921 0.7414970
# retrieve the AUC for the performance object:
perf.auc()
0.7414970113257631
# retrieve the AUC for both the training and validation data:
airlines_gbm.auc(train=True, valid=True, xval=False)
{u'train': 0.7857920674306693, u'valid': 0.7414970113257631}
AUCPR (Area Under the Precision-Recall Curve)¶
This model metric is used to evaluate how well a binary classification model is able to distinguish between precision recall pairs or points. These values are obtained using different thresholds on a probabilistic or other continuous-output classifier. AUCPR is an average of the precision-recall weighted by the probability of a given threshold.
The main difference between AUC and AUCPR is that AUC calculates the area under the ROC curve and AUCPR calculates the area under the Precision Recall curve. The Precision Recall curve does not care about True Negatives. For imbalanced data, a large quantity of True Negatives usually overshadows the effects of changes in other metrics like False Positives. The AUCPR will be much more sensitive to True Positives, False Positives, and False Negatives than AUC. As such, AUCPR is recommended over AUC for highly imbalanced data.
Note: The metric function of AUCPR only runs with command model.pr_auc. This is different than the stopping_metric which can be set equal to “AUCPR”.
Example
Using the previous example, run the following to retrieve the AUCPR.
# retrieve the AUCPR for the performance object:
h2o.pr_auc(perf)
[1] 0.7609887
# retrieve the AUCPR for both the training and validation data:
h2o.pr_auc(airlines.gbm, train=TRUE, valid=TRUE, xval=FALSE)
train valid
0.8019599 0.7609887
# retrieve the AUCPR for the performance object:
perf.pr_auc()
0.7609887253334723
# retrieve the AUCPR for both the training and validation data:
airlines_gbm.pr_auc(train=True, valid=True, xval=False)
{u'train': 0.801959918132391, u'valid': 0.7609887253334723}
Metric Best Practices - Regression¶
When deciding which metric to use in a regression problem, some main questions to ask are:
- Do you want your metric sensitive to outliers?
- What unit should the metric be in?
Sensitive to Outliers¶
Certain metrics are more sensitive to outliers. When a metric is sensitive to outliers, it means that it is important that the model predictions are never “very” wrong. For example, let’s say we have an experiment predicting number of days until an event. The graph below shows the absolute error in our predictions.
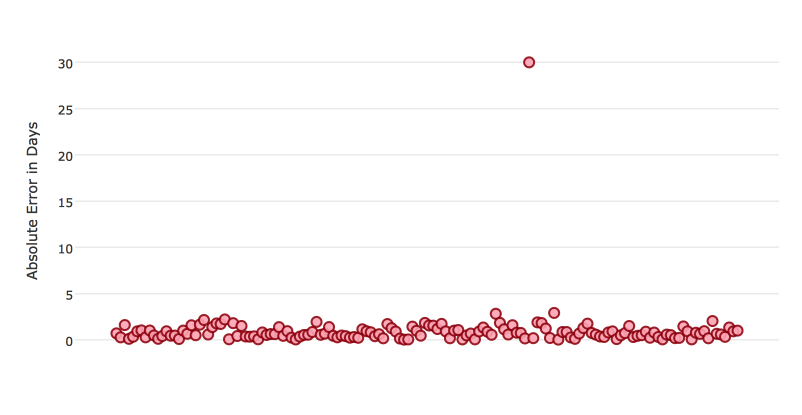
Usually our model is very good. We have an absolute error less than 1 day about 70% of the time. There is one instance, however, where our model did very poorly. We have one prediction that was 30 days off.
Instances like this will more heavily penalize metrics that are sensitive to outliers. If you do not care about these outliers in poor performance as long as you typically have a very accurate prediction, then you would want to select a metric that is robust to outliers. You can see this reflected in the behavior of the metrics: MSE and RMSE.
| MSE | RMSE | |
|---|---|---|
| Outlier | 0.99 | 2.64 |
| No Outlier | 0.80 | 1.0 |
Calculating the RMSE and MSE on our error data, the RMSE is more than twice as large as the MSE because RMSE is sensitive to outliers. If you remove the one outlier record from our calculation, RMSE drops down significantly.
Performance Units¶
Different metrics will show the performance of your model in different units. Let’s continue with our example where our target is to predict the number of days until an event. Some possible performance units are:
- Same as target: The unit of the metric is in days
- ex: MAE = 5 means the model predictions are off by 5 days on average
- Percent of target: The unit of the metric is the percent of days
- ex: MAPE = 10% means the model predictions are off by 10 percent on average
- Square of target: The unit of the metric is in days squared
- ex: MSE = 25 means the model predictions are off by 5 days on average (square root of 25 = 5)
Comparison¶
| Metric | Units | Sensitive to Outliers | Tip |
|---|---|---|---|
| R2 | scaled between 0 and 1 | No | use when you want performance scaled between 0 and 1 |
| MSE | square of target | Yes | |
| RMSE | same as target | Yes | |
| RMSLE | log of target | Yes | |
| RMSPE | percent of target | Yes | use when target values are across different scales target values are across differ ent scales |
| MAE | same as target | No | |
| MAPE | percent of target | No | use when target values are across different scales |
| SMAPE | percent of target divided by 2 | No | use when target values are close to 0 |
Metric Best Practices - Classification¶
When deciding which metric to use in a classification problem some main questions to ask are:
- Do you want the metric to evaluate the predicted probabilities or the classes that those probabilities can be converted to?
- Is your data imbalanced?
Does the Metric Evaluate Probabilities or Classes?¶
The final output of a model is a predicted probability that a record is in a particular class. The metric you choose will either evaluate how accurate the probability is or how accurate the assigned class is from that probability.
Choosing this depends on the use of the model. Do you want to use the probabilities, or do you want to convert those probabilities into classes? For example, if you are predicting whether a customer will churn, you can take the predicted probabilities and turn them into classes - customers who will churn vs customers who won’t churn. If you are predicting the expected loss of revenue, you will instead use the predicted probabilities (predicted probability of churn * value of customer).
If your use case requires a class assigned to each record, you will want to select a metric that evaluates the model’s performance based on how well it classifies the records. If your use case will use the probabilities, you will want to select a metric that evaluates the model’s performance based on the predicted probability.
Is the Metric Robust to Imbalanced Data?¶
For certain use cases, positive classes may be very rare. In these instances, some metrics can be misleading. For example, if you have a use case where 99% of the records have Class = No, then a model that always predicts No will have 99% accuracy.
For these use cases, it is best to select a metric that does not include True Negatives or considers relative size of the True Negatives like AUCPR or MCC.
Metric Comparison¶
| Metric | Evaluation Based On | Tip |
|---|---|---|
| MCC | Class | good for imbalanced data |
| F1 | Class | |
| F0.5 | Class | good when you want to give more weight to precision |
| F2 | Class | good when you want to give more weight to recall |
| Accuracy | Class | highly interpretable |
| Logloss | Probability | |
| AUC | Class | |
| AUCPR | Class | good for imbalanced data |
Stopping Model Metrics¶
Stopping metric parameters are specified in conjunction with a stopping tolerance and a number of stopping rounds. A metric specified with the stopping_metric option specifies the metric to consider when early stopping is specified.
Misclassification¶
This parameter specifies that a model must improve its misclassification rate by a given amount (specified by the stopping_tolerance parameter) in order to continue iterating. The misclassification rate is the number of observations incorrectly classified divided by the total number of observations.
Examples:
# import the airlines dataset:
from h2o.estimators import H2OGradientBoostingEstimator
airlines <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
# set the factors:
airlines["Year"] <- as.factor(airlines["Year"])
airlines["Month"] <- as.factor(airlines["Month"])
airlines["DayOfWeek"] <- as.factor(airlines["DayOfWeek"])
airlines["Cancelled"] <- as.factor(airlines["Cancelled"])
airlines['FlightNum'] <- as.factor(airlines['FlightNum'])
# set the predictors and response columns:
predictors <- c("Origin", "Dest", "Year", "UniqueCarrier",
"DayOfWeek", "Month", "Distance", "FlightNum")
response <- "IsDepDelayed"
# split the training and validation sets:
airlines.splits <- h2o.splitFrame(data = airlines, ratios = .8, seed = 1234)
train <- airlines.splits[[1]]
valid <- airlines.splits[[2]]
# build and train the model using the misclassification stopping metric:
airlines.gbm <- h2o.gbm(x = predictors, y = response,
training_frame = train, validation_frame = valid,
stopping_metric = "misclassification", stopping_rounds = 3,
stopping_tolerance = 1e-2, seed = 1234)
# retrieve the auc value:
h2o.auc(airlines.gbm, valid = TRUE)
# import H2OGradientBoostingEstimator and the airlines dataset:
from h2o.estimators import H2OGradientBoostingEstimator
airlines= h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
# set the factors:
airlines["Year"]= airlines["Year"].asfactor()
airlines["Month"]= airlines["Month"].asfactor()
airlines["DayOfWeek"] = airlines["DayOfWeek"].asfactor()
airlines["Cancelled"] = airlines["Cancelled"].asfactor()
airlines['FlightNum'] = airlines['FlightNum'].asfactor()
# set the predictors and response columns:
predictors = ["Origin", "Dest", "Year", "UniqueCarrier",
"DayOfWeek", "Month", "Distance", "FlightNum"]
response = "IsDepDelayed"
# split the training and validation sets:
train, valid= airlines.split_frame(ratios = [.8], seed = 1234)
# build and train the model using the misclassification stopping metric:
airlines_gbm = H2OGradientBoostingEstimator(stopping_metric = "misclassification",
stopping_rounds = 3,
stopping_tolerance = 1e-2,
seed = 1234)
airlines_gbm.train(x = predictors, y = response,
training_frame = train, validation_frame = valid)
# retrieve the auc value:
airlines_gbm.auc(valid=True)
Lift Top Group¶
This parameter specifies that a model must improve its lift within the top 1% of the training data. To calculate the lift, H2O sorts each observation from highest to lowest predicted value. The top group or top 1% corresponds to the observations with the highest predicted values. Lift is the ratio of correctly classified positive observations (rows with a positive target) to the total number of positive observations within a group
Examples:
# import the airlines dataset:
airlines <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
# set the factors:
airlines["Year"] <- as.factor(airlines["Year"])
airlines["Month"] <- as.factor(airlines["Month"])
airlines["DayOfWeek"] <- as.factor(airlines["DayOfWeek"])
airlines["Cancelled"] <- as.factor(airlines["Cancelled"])
airlines['FlightNum'] <- as.factor(airlines['FlightNum'])
# set the predictors and response columns:
predictors <- c("Origin", "Dest", "Year", "UniqueCarrier",
"DayOfWeek", "Month", "Distance", "FlightNum")
response <- "IsDepDelayed"
# split the training and validation sets:
airlines.splits <- h2o.splitFrame(data = airlines, ratios = .8, seed = 1234)
train <- airlines.splits[[1]]
valid <- airlines.splits[[2]]
# build and train the model using the lift_top_group stopping metric:
airlines.gbm <- h2o.gbm(x = predictors, y = response,
training_frame = train, validation_frame = valid,
stopping_metric = "lift_top_group", stopping_rounds = 3,
stopping_tolerance = 1e-2, seed = 1234)
# retrieve the auc value:
h2o.auc(airlines.gbm, valid = TRUE)
# import H2OGradientBoostingEstimator and the airlines dataset:
from h2o.estimators import H2OGradientBoostingEstimator
airlines= h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
# set the factors:
airlines["Year"]= airlines["Year"].asfactor()
airlines["Month"]= airlines["Month"].asfactor()
airlines["DayOfWeek"] = airlines["DayOfWeek"].asfactor()
airlines["Cancelled"] = airlines["Cancelled"].asfactor()
airlines['FlightNum'] = airlines['FlightNum'].asfactor()
# set the predictors and response columns:
predictors = ["Origin", "Dest", "Year", "UniqueCarrier",
"DayOfWeek", "Month", "Distance", "FlightNum"]
response = "IsDepDelayed"
# split the training and validation sets:
train, valid= airlines.split_frame(ratios = [.8], seed = 1234)
# build and train the model using the lifttopgroup stopping metric:
airlines_gbm = H2OGradientBoostingEstimator(stopping_metric = "lifttopgroup",
stopping_rounds = 3,
stopping_tolerance = 1e-2,
seed = 1234)
airlines_gbm.train(x = predictors, y = response,
training_frame = train, validation_frame = valid)
# retrieve the auc value:
airlines_gbm.auc(valid=True)
Deviance¶
The model will stop building if the deviance fails to continue to improve. Deviance is computed as follows:
Loss = Quadratic -> MSE==Deviance For Absolute/Laplace or Huber -> MSE != Deviance
Examples:
# import the cars dataset:
cars <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the predictors and response columns:
predictors <- c("economy","cylinders","displacement","power","weight")
response = "acceleration"
#split the training and validation sets:
p.sid <- h2o.runif(cars, seed=1234)
train <- h2o.assign(cars[p.sid > .2, ], "train")
test <- h2o.assign(cars[p.sid <= .2, ], "test")
# build and train the model using the deviance stopping metric:
cars_gbm <- h2o.gbm(x=predictors, y=repsonse,
training_frame=train, validation_frame=test,
stopping_metric = "deviance", stopping_rounds = 3,
stopping_tolerance = 1e-2, seed = 1234)
# retrieve the mse value:
h2o.mse(cars_gbm, valid=TRUE)
# import H2OGradientBoostingEstimator and the cars dataset:
from h2o.estimators import H2OGradientBoostingEstimator
cars = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the predictors and response columns:
predictors = ["economy","cylinders","displacement","power","weight"]
response = "acceleration"
# split the training and validation sets:
train, valid = cars.split_frame(ratios=[.8],seed=1234)
# build and train the model using the deviance stopping metric:
cars_gbm = H2OGradientBoostingEstimator(stopping_metric = "deviance",
stopping_rounds = 3,
stopping_tolerance = 1e-2,
seed = 1234)
cars_gbm.train(x=predictors, y=response,
training_frame=train, validation_frame=valid)
# retrieve the mse value:
cars_gbm.mse(valid=True)
Mean-Per-Class-Error¶
The model will stop building after the mean-per-class error rate fails to improve.
Examples:
# import the cars dataset:
cars <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the predictors and response columns:
predictors <- c("economy","cylinders","displacement","power","weight")
response = "acceleration"
#split the training and validation sets:
p.sid <- h2o.runif(cars, seed=1234)
train <- h2o.assign(cars[p.sid > .2, ], "train")
test <- h2o.assign(cars[p.sid <= .2, ], "test")
# build and train the model using the mean_per_class_error stopping metric:
cars_gbm <- h2o.gbm(x=predictors, y=repsonse,
training_frame=train, validation_frame=test,
stopping_metric = "mean_per_class_error", stopping_rounds = 3,
stopping_tolerance = 1e-2, seed = 1234)
# retrieve the mse value:
h2o.mse(cars_gbm, valid=TRUE)
# import H2OGradientBoostingEstimator and the cars dataset:
from h2o.estimators import H2OGradientBoostingEstimator
cars = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the predictors and response columns:
predictors = ["economy","cylinders","displacement","power","weight"]
response = "acceleration"
# split the training and validation sets:
train, valid = cars.split_frame(ratios=[.8],seed=1234)
# build and train the model using the meanperclasserror stopping metric:
cars_gbm = H2OGradientBoostingEstimator(stopping_metric = "meanperclasserror",
stopping_rounds = 3,
stopping_tolerance = 1e-2,
seed = 1234)
cars_gbm.train(x=predictors, y=repsonse,
training_frame=train, validation_frame=valid)
# retrieve the mse value:
cars_gbm.mse(valid=True)
In addition to the above options, Logloss, MSE, RMSE, MAE, RMSLE, and AUC can also be used as the stopping metric.
Model Performance Graphs¶
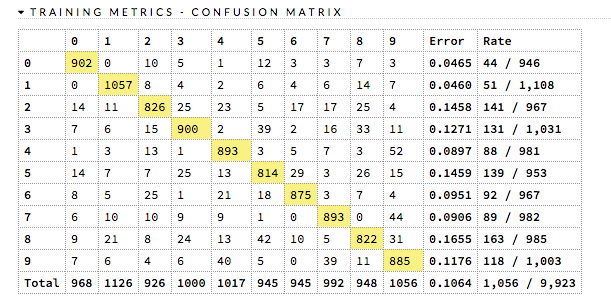
Confusion Matrix¶
A confusion matrix is a table depicting performance of algorithm in terms of false positives, false negatives, true positives, and true negatives. In H2O, the actual results display in the columns and the predictions display in the rows; correct predictions are highlighted in yellow. In the example below, 0 was predicted correctly 902 times, while 8 was predicted correctly 822 times and 0 was predicted as 4 once.
Examples:
# import the cars dataset:
cars <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the factor
cars["cylinders"] = as.factor(cars["cylinders"])
# split the training and validation sets:
cars.splits <- h2o.splitFrame(data = cars, ratio = .8, seed = 1234)
train <- cars.splits[[1]]
valid <- cars.splits[[2]]
# set the predictors columns, response column, and distribution type:
predictors <- c("displacement","power","weight","acceleration","year")
response <- "cylinders"
distribution <- "multinomial"
# build and train the model:
cars_gbm <- h2o.gbm(x=predictors, y=response,
training_frame=train, validation_frame = valid,
nfolds=3, distribution=distribution)
# build the confusion matrix:
h2o.confusionMatrix(cars_gbm)
# import H2OGradientBoostingEstimator and the cars dataset:
cars = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the factor:
cars["cylinders"] = cars["cylinders"].asfactor()
# split the training and validation sets:
r = cars[0].runif()
train = cars[r > .2]
valid = cars[r <= .2]
# set the predictors columns, response column, and distribution type:
predictors = ["displacement","power","weight","acceleration","year"]
response_col = "cylinders"
distribution = "multinomial"
# build and train the model:
gbm = H2OGradientBoostingEstimator(nfolds = 3, distribution = distribution)
gbm.train(x=predictors, y=response_col, training_frame=train, validation_frame=valid)
# build the confusion matrix:
gbm.confusion_matrix(train)
Variable Importances¶
Variable importances represent the statistical significance of each variable in the data in terms of its affect on the model. Variables are listed in order of most to least importance. The percentage values represent the percentage of importance across all variables, scaled to 100%. The method of computing each variable’s importance depends on the algorithm. More information is available in the Variable Importance section.
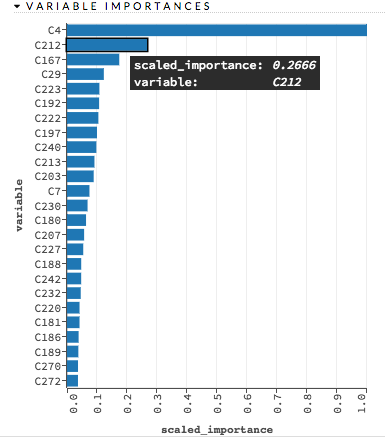
Examples:
# import the prostate dataset:
prostate <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factor:
prostate[,2] <- as.factor(prostate[,2])
# split the training and validation sets:
pros.split <- h2o.splitFrame(data = prostate, ratio = .8, seed = 1234)
train <- pros.split[[1]]
valid <- pros.split[[2]]
# build and train the model:
pros_gbm <- h2o.gbm(x = 3:9, y = 2,
training_frame = train,
validation_frame = valid,
distribution = "bernoulli")
# build the variable importances plot:
h2o.varimp_plot(pros_gbm)
# import H2OGradientBoostingEstimator and the prostate dataset:
from h2o.estimators import H2OGradientBoostingEstimator
pros = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factors:
pros[1] = pros[1].asfactor()
pros[3] = pros[3].asfactor()
pros[4] = pros[4].asfactor()
pros[5] = pros[5].asfactor()
pros[8] = pros[8].asfactor()
# split the training and validation sets:
r = pros[1].runif()
train = pros[r > .2]
valid = pros[r <= .2]
# set the predictors and response columns:
predictors = ["AGE","RACE","DPROS","DCAPS","PSA","VOL","GLEASON"]
response = "CAPSULE"
# build and train the model:
pros_gbm = H2OGradientBoostingEstimator(nfolds=2)
pros_gbm.train(x = predictors, y = response,
training_frame = train,
validation_frame = valid)
# build the variable importances plot:
pros_gbm.varimp_plot()
ROC Curve¶
A ROC Curve is a graph that represents the ratio of true positives to false positives. (For more information, refer to the Linear Digressions podcast describing ROC Curves.) To view a specific threshold, select a value from the drop-down Threshold list. To view any of the following details, select it from the drop-down Criterion list:
- Max f1
- Max f2
- Max f0point5
- Max accuracy
- Max precision
- Max absolute MCC (the threshold that maximizes the absolute Matthew’s Correlation Coefficient)
- Max min per class accuracy
The lower-left side of the graph represents less tolerance for false positives while the upper-right represents more tolerance for false positives. Ideally, a highly accurate ROC resembles the following example.
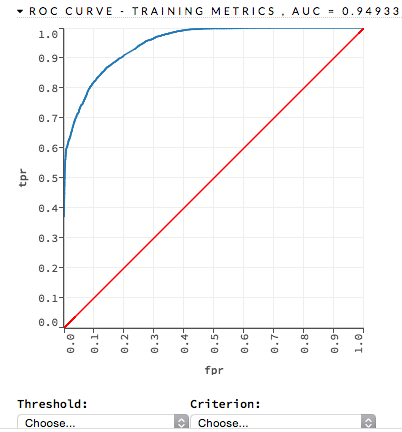
Examples:
# import the prostate dataset:
pros <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factors:
pros[,2] <- as.factor(pros[,2])
pros[,4] <- as.factor(pros[,4])
pros[,5] <- as.factor(pros[,5])
pros[,6] <- as.factor(pros[,6])
pros[,9] <- as.factor(pros[,9])
# split the training and validation sets:
p.sid <- h2o.runif(pros, seed=1234)
pros.train <- h2o.assign(pros[p.sid > .2, ], "pros.train")
pros.test <- h2o.assign(pros[p.sid <= .2, ], "pros.test")
# build and train the model:
pros_gbm <- h2o.gbm(x = 3:9, y = 2,
training_frame = pros.train,
validation_frame = pros.test,
nfolds = 2)
# build the roc curve:
perf <- h2o.performance(pros_gbm, pros)
plot(perf, type="roc")
# import H2OGradientBoostingEstimator and the prostate dataset:
from h2o.estimators import H2OGradientBoostingEstimator
pros = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factors:
pros[1] = pros[1].asfactor()
pros[3] = pros[3].asfactor()
pros[4] = pros[4].asfactor()
pros[5] = pros[5].asfactor()
pros[8] = pros[8].asfactor()
# set the predictors and response columns:
predictors = ["AGE","RACE","DPROS","DCAPS","PSA","VOL","GLEASON"]
response = "CAPSULE"
# split the training and validation sets:
r = pros[1].runif()
train = pros[r > .2]
valid = pros[r <= .2]
# build and train the model:
pros_gbm = H2OGradientBoostingEstimator(nfolds=2)
pros_gbm.train(x = predictors, y = response, training_frame = pros)
# build the roc curve:
perf = pros_gbm.model_performance(pros)
perf.plot(type = "roc")
Hit Ratio¶
The hit ratio is a table representing the number of times that the prediction was correct out of the total number of predictions.
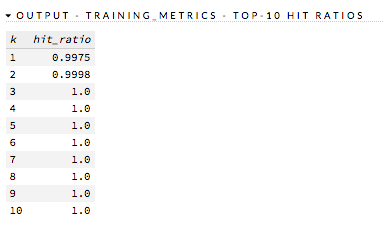
Examples:
# import the cars dataset:
cars <- h2o.importFile("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the factor:
cars["cylinders"] = as.factor(cars["cylinders"])
# split the training and validation sets:
cars.splits <- h2o.splitFrame(data = cars, ratio = .8, seed = 1234)
train <- cars.splits[[1]]
valid <- cars.splits[[2]
# set the predictors columns, response column, and distribution type:
predictors <- c("displacement","power","weight","acceleration","year")
response <- "cylinders"
distribution <- "multinomial"
# build and train model:
cars_gbm <- h2o.gbm(x=predictors, y=response,
training_frame=train, validation_frame = valid,
nfolds=3, distribution=distribution)
# build the hit ratio table:
gbm_hit <- h2o.hit_ratio_table(cars_gbm, train = FALSE, valid = FALSE)
gbm_hit
# import H2OGradientBoostingEstimator and the cars dataset:
from h2o.estimators import H2OGradientBoostingEstimator
cars = h2o.import_file("https://s3.amazonaws.com/h2o-public-test-data/smalldata/junit/cars_20mpg.csv")
# set the factor:
cars["cylinders"] = cars["cylinders"].asfactor()
# split the training and validation sets:
r = cars[0].runif()
train = cars[r > .2]
valid = cars[r <= .2]
# set the predictors columns, repsonse column, and distribution type:
predictors = ["displacement","power","weight","acceleration","year"]
response_col = "cylinders"
distribution = "multinomial"
# build and train the model:
gbm = H2OGradientBoostingEstimator(nfolds = 3, distribution = distribution)
gbm.train(x=predictors, y=response_col, training_frame=train, validation_frame=valid)
# build the hit ratio table:
gbm_hit = gbm.hit_ratio_table(valid=True)
gbm_hit.show()
Standardized Coefficient Magnitudes¶
This chart represents the relationship of a specific feature to the response variable. Coefficients can be positive (orange) or negative (blue). A positive coefficient indicates a positive relationship between the feature and the response, where an increase in the feature corresponds with an increase in the response, while a negative coefficient represents a negative relationship between the feature and the response where an increase in the feature corresponds with a decrease in the response (or vice versa).
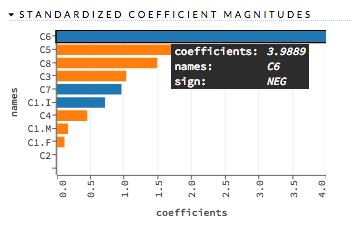
Examples:
# import the prostate dataset:
prostate <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factor:
prostate[,2] <- as.factor(prostate[,2])
# set the predictors and response columns:
response <- "CAPSULE"
predictors <- c("AGE","RACE","PSA","DCAPS")
# build and train the model:
pros_glm <- h2o.glm(x = predictors, y = response,
training_frame = prostate,
family = "binomial", nfolds = 0,
alpha = 0.5, lambda_search = FALSE)
# build the standardized coefficient magnitudes plot:
h2o.std_coef_plot(pros_glm)
# import H2OGeneralizedLinearEstimator and the prostate dataset:
from h2o.estimators import H2OGeneralizedLinearEstimator
prostate = h2o.import_file("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factor:
prostate["CAPSULE"] = prostate["CAPSULE"].asfactor()
# set the predictors and response columns:
response = "CAPSULE"
predictors = ["AGE","RACE","PSA","DCAPS"]
# build and train the model:
glm = H2OGeneralizedLinearEstimator(nfolds = 5, alpha = 0.5,
lambda_search = False,
family = "binomial")
glm.train(x=predictors, y=response, training_frame=prostate)
# build the standardized coefficient magnitudes plot:
glm.std_coef_plot()
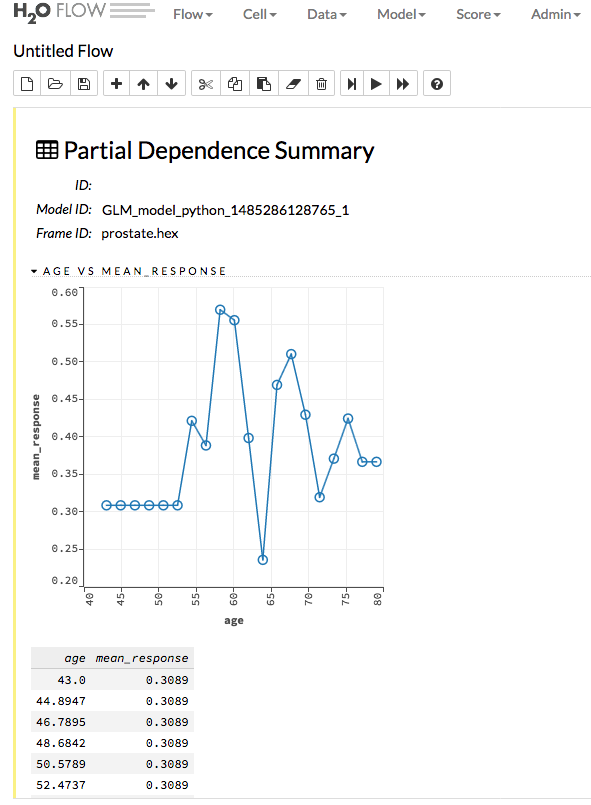
Partial Dependence Plots¶
This plot provides a graphical representation of the marginal effect of a variable on the class probability (classification) or response (regression). Note that this is only available for models that include only numerical values.
The partial dependence of a given feature \(X_j\) is the average of the response function \(g\), where all the components of \(X_j\) are set to \(x_j\) \((X_j = {[x{^{(0)}_j},...,x{^{(N-1)}_j}]}^T)\)
Thus, the one-dimensional partial dependence of function \(g\) on \(X_j\) is the marginal expectation:
Notes:
- The partial dependence of a given feature is \(Xj\) (where \(j\) is the column index)
- You can also change the equation to sum from 1 to N instead of 0 to N-1
- Use the
col_pairs_2dpdpoption along with a list containing pairs of column names to generate 2D partial dependence plots
Examples:
# import the prostate dataset:
prostate <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factors:
prostate[, "CAPSULE"] <- as.factor(prostate[, "CAPSULE"])
prostate[, "RACE"] <- as.factor(prostate[, "RACE"])
# build and train the model:
pros_gbm <- h2o.gbm(x = c("AGE","RACE"), y = "CAPSULE",
training_frame = prostate,
ntrees = 10, max_depth = 5,
learn_rate = 0.1)
# build the partial dependence plot:
h2o.partialPlot(object = pros_gbm, data = prostate, cols = c("AGE","RACE"))
# import H2OGradiantBoostingEstimator and the prostate dataset:
from h2o.estimators import H2OGradientBoostingEstimator
prostate = h2o.import_file("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
# set the factors:
prostate["CAPSULE"] = prostate["CAPSULE"].asfactor()
prostate["RACE"] = prostate["RACE"].asfactor()
# set the predictors and response columns:
predictors = ["AGE","RACE"]
response = "CAPSULE"
# build and train the model:
pros_gbm = H2OGradientBoostingEstimator(ntrees = 10, max_depth = 5, learn_rate = 0.1)
pros_gbm.train(x = predictors, y = response, training_frame = prostate)
#build the partial dependence plot:
pros_gbm.partial_plot(data = prostate, cols = ["AGE","RACE"], server=True, plot = True)
Prediction¶
With H2O-3, you can generate predictions for a model based on samples in a test set using h2o.predict() or predict(). This can be accomplished in memory or using MOJOs/POJOs.
Note: MOJO/POJO predict cannot parse columns enclosed in double quotes (for example, “”2””).
For classification problems, predicted probabilities and labels are compared against known results. (Note that for binary models, labels are based on the maximum F1 threshold from the model object.) For regression problems, predicted regression targets are compared against testing targets and typical error metrics.
In-Memory Prediction¶
This section provides examples of performing predictions in Python and R. Refer to the Predictions topic in the Flow chapter to view an example of how to predict in Flow.
library(h2o)
h2o.init()
# Import the prostate dataset
prostate.hex <- h2o.importFile(path = "https://raw.github.com/h2oai/h2o/master/smalldata/logreg/prostate.csv",
destination_frame = "prostate.hex")
# Split dataset giving the training dataset 75% of the data
prostate.split <- h2o.splitFrame(data=prostate.hex, ratios=0.75)
# Create a training set from the 1st dataset in the split
prostate.train <- prostate.split[[1]]
# Create a testing set from the 2nd dataset in the split
prostate.test <- prostate.split[[2]]
# Convert the response column to a factor
prostate.train$CAPSULE <- as.factor(prostate.train$CAPSULE)
# Build a GBM model
model <- h2o.gbm(y="CAPSULE",
x=c("AGE", "RACE", "PSA", "GLEASON"),
training_frame=prostate.train,
distribution="bernoulli",
ntrees=100,
max_depth=4,
learn_rate=0.1)
# Predict using the GBM model and the testing dataset
pred <- h2o.predict(object=model, newdata=prostate.test)
pred
predict p0 p1
1 0 0.7414373 0.25856274
2 1 0.3114293 0.68857073
3 0 0.9852284 0.01477161
4 0 0.6647902 0.33520975
5 0 0.6075046 0.39249538
6 1 0.4065468 0.59345323
[88 rows x 3 columns]
# View a summary of the prediction with a probability of TRUE
summary(pred$p1, exact_quantiles=TRUE)
p1
Min. :0.008925
1st Qu.:0.160050
Median :0.350236
Mean :0.451507
3rd Qu.:0.818486
Max. :0.99040
import h2o
from h2o.estimators.gbm import H2OGradientBoostingEstimator
h2o.init()
# Import the prostate dataset
h2o_df = h2o.import_file("https://raw.github.com/h2oai/h2o/master/smalldata/logreg/prostate.csv")
# Split the data into Train/Test/Validation with Train having 70% and test and validation 15% each
train,test,valid = h2o_df.split_frame(ratios=[.7, .15])
# Convert the response column to a factor
h2o_df["CAPSULE"] = h2o_df["CAPSULE"].asfactor()
# Generate a GBM model using the training dataset
model = H2OGradientBoostingEstimator(distribution="bernoulli",
ntrees=100,
max_depth=4,
learn_rate=0.1)
model.train(y="CAPSULE", x=["AGE","RACE","PSA","GLEASON"],training_frame=h2o_df)
# Predict using the GBM model and the testing dataset
predict = model.predict(test)
# View a summary of the prediction
predict.head()
predict p0 p1
--------- -------- --------
0 0.8993 0.1007
1 0.168391 0.831609
1 0.166067 0.833933
1 0.327212 0.672788
1 0.25991 0.74009
0 0.758978 0.241022
0 0.540797 0.459203
0 0.838489 0.161511
0 0.704853 0.295147
0 0.642381 0.357619
[10 rows x 3 columns]
Predicting Leaf Node Assignment¶
For tree-based models, the h2o.predict_leaf_node_assignment() function predicts the leaf node assignment on an H2O model.
This function predicts against a test frame. For every row in the test frame, this function returns the leaf placements of the row in all the trees in the model. An optional Type can also be specified to define the placements. Placements can be represented either by paths to the leaf nodes from the tree root (Path - default) or by H2O’s internal identifiers (Node_ID). The order of the rows in the results is the same as the order in which the data was loaded.
This function returns an H2OFrame object with categorical leaf assignment identifiers for each tree in the model.
Using the previous example, run the following to predict the leaf node assignments:
# Predict the leaf node assigment using the GBM model and test data.
# Predict based on the path from the root node of the tree.
predict_lna <- h2o.predict_leaf_node_assignment(model, prostate.test)
# View a summary of the leaf node assignment prediction
summary(predict_lna$T1.C1, exact_quantiles=TRUE)
T1.C1
RRLR:15
RRR :13
LLLR:12
LLLL:11
LLRR: 8
LLRL: 6
# Predict the leaf node assigment using the GBM model and test data.
# Predict based on the path from the root node of the tree.
predict_lna = model.predict_leaf_node_assignment(test, "Path")
Predict Contributions¶
In H2O-3, each returned H2OFrame has a specific shape (#rows, #features + 1). This includes a feature contribution column for each input feature, with the last column being the model bias (same value for each row). The sum of the feature contributions and the bias term is equal to the raw prediction of the model. Raw prediction of tree-based model is the sum of the predictions of the individual trees before the inverse link function is applied to get the actual prediction. For Gaussian distribution, the sum of the contributions is equal to the model prediction.
H2O-3 supports TreeSHAP for DRF, GBM, and XGBoost. For these problems, the predict_contributions returns a new H2OFrame with the predicted feature contributions - SHAP (SHapley Additive exPlanation) values on an H2O model. If you have SHAP installed, then raphical representations can be retrieved in Python using SHAP functions. (Note that retrieving graphs via R is not yet supported.) An .ipynb demo showing this example is also available here.
Note: Multinomial classification models are currently not supported.
# Predict the contributions using the GBM model and test data.
contributions <- h2o.predict_contributions(model, prostate.test)
contributions
AGE RACE PSA GLEASON BiasTerm
--------- ---------- --------- --------- ----------
-0.3929753 0.02188157 0.3530045 0.5453218 -0.6589417
-0.6489378 -0.24417394 1.0434356 0.7937416 -0.6589417
0.3244801 -0.23901901 0.9877144 1.0463049 -0.6589417
0.9402978 -0.33412665 2.0499718 1.0571480 -0.6589417
-0.7762397 0.03393304 0.1952782 1.8620299 -0.6589417
0.5900557 0.03899451 0.6708371 -1.2606093 -0.6589417
[95 rows x 5 columns]
# Predict the contributions using the GBM model and test data.
contributions = model.predict_contributions(test)
contributions
AGE RACE PSA GLEASON BiasTerm
----------- ---------- --------- --------- ----------
-0.414587 0.0263119 -0.120703 0.407889 -0.581522
0.0913486 0.0250697 -0.746584 1.16642 -0.581522
0.565866 0.0603216 2.51301 0.739406 -0.581522
-0.670981 0.0210115 0.164873 -2.03487 -0.581522
-0.398603 0.0255295 -0.494069 0.537647 -0.581522
0.00915739 0.0458912 0.557667 -0.262171 -0.581522
-0.199497 -0.265438 2.18964 2.89974 -0.581522
-0.137073 0.0271401 -1.00939 1.47302 -0.581522
0.440857 0.0407717 -0.574498 -0.537758 -0.581522
-0.901466 0.0216657 0.453894 -2.39536 -0.581522
[58 rows x 5 columns]
# Import required packages for running SHAP commands
import shap
# Load JS visualization code
shap.initjs()
# Convert the H2OFrame to use with SHAP's visualization functions
contributions_matrix = contributions.as_data_frame().as_matrix()
# Calculate SHAP values for all features
shap_values = contributions_matrix[:,0:4]
# Expected values is the last returned column
expected_value = contributions_matrix[:,4].min()
# Visualize the training set predictions
X=["AGE","RACE","PSA","GLEASON"]
shap.force_plot(expected_value, shap_values, X)
# Summarize the effects of all the features
shap.summary_plot(shap_values, X)
# View the same summary as a bar chart
shap.summary_plot(shap_values, X, plot_type="bar")
Predict Stage Probabilities¶
Use the staged_predict_proba function to predict class probabilities at each stage of an H2O Model. Note that this can only be used with GBM.
Using the previous example, run the following to predict probabilities at each stage in the model:
# Predict the class probabilities using the GBM model and test data.
staged_predict_proba <- h2o.staged_predict_proba(model, prostate.test)
# Predict the class probabilities using the GBM model and test data.
staged_predict_proba = model.staged_predict_proba(test)
Prediction Threshold¶
For classification problems, when running h2o.predict() or .predict(), the prediction threshold is selected as follows:
- If you only have training data, the max F1 threshold comes from the train data model.
- If you train a model with training and validation data, the max F1 threshold comes from the validation data model.
- If you train a model with training data and set the
nfoldsparameter, the Max F1 threshold from the training data model metrics is used. - If you train a model with the train data and validation data and also set the
nfoldsparameter, the Max F1 threshold from the validation data model metrics is used.
Predict Feature Frequency¶
Use the feature_frequencies function to retrieve the number of times a feature was used on a prediction path in a tree model. This option is only available in GBM, DRF, and IF.
Using the previous example, run the following to the find frequency of each feature in the prediction path of the model:
# Retrieve the number of occurrences of each feature for given observations
# on their respective paths in a tree ensemble model
feature_frequencies <- h2o.feature_frequencies(model, prostate.train)
feature_frequencies
AGE RACE PSA GLEASON
98 8 199 46
114 6 238 42
103 9 227 57
94 13 183 53
103 9 225 57
102 5 238 36
[275 rows x 4 columns]
# Retrieve the number of occurrences of each feature for given observations
# on their respective paths in a tree ensemble model
feature_frequencies = model.feature_frequencies(train)
feature_frequencies
AGE RACE PSA GLEASON
----- ------ ----- ---------
109 10 197 68
109 3 220 64
101 11 222 66
106 6 188 65
90 1 199 61
130 7 194 65
103 3 217 66
103 11 203 65
102 3 218 66
112 6 203 64
[273 rows x 4 columns]
Predict using MOJOs¶
An end-to-end example from building a model through predictions using MOJOs is available in the MOJO Quick Start topic.
Predict using POJOs¶
An end-to-end example from building a model through predictions using POJOs is available in the POJO Quick Start topic.