Productionizing H2O¶
About POJOs and MOJOs¶
H2O allows you to convert the models you have built to either a Plain Old Java Object (POJO) or a Model ObJect, Optimized (MOJO).
H2O-generated MOJO and POJO models are intended to be easily embeddable in any Java environment. The only compilation and runtime dependency for a generated model is the h2o-genmodel.jar file produced as the build output of these packages. This file is a library that supports scoring. For POJOs, it contains the base classes from which the POJO is derived from. (You can see “extends GenModel” in a POJO class. The GenModel class is part of this library.) For MOJOs, it also contains the required readers and interpreters. The h2o-genmodel.jar file is required when POJO/MOJO models are deployed to production.
Users can refer to the Quick Start topics that follow for more information about generating POJOs and MOJOs.
Developers can refer to the the POJO and MOJO Model Javadoc.
MOJO Quick Start¶
This section describes how to build and implement a MOJO (Model Object, Optimized) to use predictive scoring. Java developers should refer to the Javadoc for more information, including packages.
What is a MOJO?¶
A MOJO (Model Object, Optimized) is an alternative to H2O’s POJO. As with POJOs, H2O allows you to convert models that you build to MOJOs, which can then be deployed for scoring in real time.
Notes:
MOJOs are supported for AutoML, Deep Learning, DRF, GBM, GLM, GLRM, K-Means, PCA, Stacked Ensembles, SVM, Word2vec, and XGBoost models.
MOJOs are only supported for encodings that are either default or
enum.MOJO predict cannot parse columns enclosed in double quotes (for example, “”2””).
Benefits of MOJOs over POJOs¶
While POJOs continue to be supported, some customers encountered issues with large POJOs not compiling. (Note that POJOs are not supported for source files larger than 1G.) MOJOs do not have a size restriction and address the size issue by taking the tree out of the POJO and using generic tree-walker code to navigate the model. The resulting executable is much smaller and faster than a POJO.
At large scale, new models are roughly 20-25 times smaller in disk space, 2-3 times faster during “hot” scoring (after JVM is able to optimize the typical execution paths), and 10-40 times faster in “cold” scoring (when JVM doesn’t know yet know the execution paths) compared to POJOs. The efficiency gains are larger the bigger the size of the model.
H2O conducted in-house testing using models with 5000 trees of depth 25. At very small scale (50 trees / 5 depth), POJOs were found to perform ≈10% faster than MOJOs for binomial and regression models, but 50% slower than MOJOs for multinomial models.
Building a MOJO¶
MOJOs are built in much the same way as POJOs. The example code below shows how to start H2O, build a model using either R or Python, and then compile and run the MOJO. This example uses GBM, but any supported algorithm can be used to build a model and run the MOJO.
The examples below describe how to start H2O and create a model using R, Python, Java, and Scala. The download_mojo() function saves the model as a zip file. You can unzip the file to view the options used to build the file along with each tree built in the model. Note that each tree file is saved as a binary file type.
Step 1: Build and Extract a Model¶
# 1. Open a terminal window and start R.
# 2. Run the following commands to build a simple GBM model.
library(h2o)
h2o.init(nthreads = -1)
path <- system.file("extdata", "prostate.csv", package="h2o")
h2o_df <- h2o.importFile(path)
h2o_df$CAPSULE <- as.factor(h2o_df$CAPSULE)
model <- h2o.gbm(y = "CAPSULE",
x = c("AGE", "RACE", "PSA", "GLEASON"),
training_frame = h2o_df,
distribution = "bernoulli",
ntrees = 100,
max_depth = 4,
learn_rate = 0.1)
# Download the MOJO and the resulting h2o-genmodel.jar file
# to a new **experiment** folder. Note that the ``h2o-genmodel.jar`` file
# is a library that supports scoring and contains the required readers
# and interpreters. This file is required when MOJO models are deployed
# to production. Be sure to specify the entire path, not just the relative path.
modelfile <- h2o.download_mojo(model, path="~/experiments/", get_genmodel_jar=TRUE)
print("Model saved to " + modelfile)
Model saved to /Users/user/GBM_model_R_1475248925871_74.zip"
# 1. Open a terminal window and start python.
# 2. Run the following commands to build a simple GBM model.
# The model, along with the **h2o-genmodel.jar** file will
# then be downloaded to an **experiment** folder.
import h2o
from h2o.estimators.gbm import H2OGradientBoostingEstimator
h2o.init()
h2o_df = h2o.load_dataset("prostate.csv")
h2o_df["CAPSULE"] = h2o_df["CAPSULE"].asfactor()
model=H2OGradientBoostingEstimator(distribution="bernoulli",
ntrees=100,
max_depth=4,
learn_rate=0.1)
model.train(y="CAPSULE",
x=["AGE","RACE","PSA","GLEASON"],
training_frame=h2o_df)
# Download the MOJO and the resulting ``h2o-genmodel.jar`` file
# to a new **experiment** folder. Note that the ``h2o-genmodel.jar`` file
# is a library that supports scoring and contains the required readers
# and interpreters. This file is required when MOJO models are deployed
# to production. Be sure to specify the entire path, not just the relative path.
modelfile = model.download_mojo(path="~/experiment/", get_genmodel_jar=True)
print("Model saved to " + modelfile)
Model saved to /Users/user/GBM_model_python_1475248925871_888.zip
// Compile the source:
javac -classpath ~/h2o/h2o-3.20.0.1/h2o.jar src/h2oDirect/h2oDirect.java
// Execute as a classfile. This also downloads the LoanStats4 demo,
// which trains a GBM model.
Erics-MBP-2:h2oDirect ericgudgion$ java -cp /Users/ericgudgion/NetBeansProjects/h2oDirect/src/:/Users/ericgudgion/h2o/h2o-3.20.0.1/h2o.jar h2oDirect.h2oDirect /Demos/Lending-Club/LoanStats4.csv
...
06-14 20:40:29.420 192.168.1.160:54321 55005 main INFO: Found XGBoost backend with library: xgboost4j_minimal
06-14 20:40:29.428 192.168.1.160:54321 55005 main INFO: Your system supports only minimal version of XGBoost (no GPUs, no multithreading)!
06-14 20:40:29.428 192.168.1.160:54321 55005 main INFO: ----- H2O started -----
06-14 20:40:29.428 192.168.1.160:54321 55005 main INFO: Build git branch: rel-wright
...
...
Starting H2O with IP 192.168.1.160:54321
Loading data from file
...
Loaded file /Demos/Lending-Club/LoanStats4.csv size 3986423 Cols:19 Rows:39029
...
Creating GBM Model
Training Model
...
Training Results
Model Metrics Type: Binomial
Description: N/A
model id: GBM_model_1529023227180_1
frame id: dataset-key
MSE: 0.11255783
RMSE: 0.3354964
AUC: 0.82892376
logloss: 0.36827797
mean_per_class_error: 0.26371866
default threshold: 0.261136531829834
...
Model AUC 0.8289237508508612
Model written out as a MOJO to file /Demos/Lending-Club/LoanStats4.csv.zip
// Save as h2oDirect.java
package h2oDirect;
import hex.tree.gbm.GBM;
import hex.tree.gbm.GBMModel;
import hex.tree.gbm.GBMModel.GBMParameters;
import java.io.FileOutputStream;
import java.io.IOException;
import java.net.InetAddress;
import water.Key;
import water.fvec.Frame;
import water.fvec.NFSFileVec;
import water.parser.ParseDataset;
import water.*;
public class h2oDirect {
/**
* @param args the command line arguments
*/
public static void main(String[] args) throws IOException {
String h2oargs = "-nthreads -1 ";
H2OApp.main(h2oargs.split(" "));
System.out.println("Starting H2O with IP "+H2O.getIpPortString());
H2O.waitForCloudSize(1, 3000);
System.out.println("Loading data from file ");
String inputfile = args[0];
NFSFileVec datafile = NFSFileVec.make(inputfile);
Frame dataframe = ParseDataset.parse(Key.make("dataset-key") , datafile._key);
System.out.println("Loaded file "+inputfile+" size "+datafile.byteSize()+" Cols:"+dataframe.numCols()+" Rows:"+dataframe.numRows());
for (int v=0; v<dataframe.numCols(); v++) {
System.out.println(dataframe.name(v)+" "+dataframe.vec(v).get_type_str());
}
int c = dataframe.find("bad_loan");
dataframe.replace(c, dataframe.vec(c).toCategoricalVec());
// drop the id and member_id columns from model
dataframe.remove(dataframe.find("id"));
dataframe.remove(dataframe.find("member_id"));
System.out.println("Creating GBM Model");
GBMParameters modelparms = new GBMParameters();
modelparms._train = dataframe._key;
modelparms._response_column = "bad_loan";
System.out.println("Training Model");
GBM model = new GBM(modelparms);
GBMModel gbm = model.trainModel().get();
System.out.println("Training Results");
System.out.println(gbm._output);
System.out.println("Model AUC "+gbm.auc());
String outputfile = inputfile+".zip";
FileOutputStream modeloutput = new FileOutputStream(outputfile);
gbm.getMojo().writeTo(modeloutput);
modeloutput.close();
System.out.println("Model written out as a MOJO to file "+outputfile);
System.out.println("H2O shutdown....");
H2O.shutdown(0);
}
}
import water.rapids.ast.prims.advmath.AstCorrelation
object RandomForestFileInput {
import water.H2O
import water.H2OApp
import water.fvec.Vec
import water.fvec.NFSFileVec
import water.fvec._
import hex.tree.drf.DRF
import hex.tree.drf.DRFModel
import hex.tree.drf.DRFModel.DRFParameters
import water.parser.ParseDataset
import water.Key
import water.Futures
import water._
import scala.io.Source
import scala.reflect._
import java.io.FileOutputStream
import java.io.FileWriter
def main(args: Array[String]): Unit = {
println("H2O Random Forest FileInput example\n")
if (args.length==0) {
println("Input file missing, please pass datafile as the first parameter")
return
}
// Start H2O instance and wait for 3 seconds for instance to complete startup
println("Starting H2O")
val h2oargs = "-nthreads -1 -quiet"
H2OApp.main(h2oargs.split(" "))
H2O.waitForCloudSize(1, 3000)
println("H2O available")
// Load datafile passed as first parameter and print the size of the file as confirmation
println("Loading data from file ")
val inputfile = args(0)
val parmsfile = args(1)
def ignore: Boolean = System.getProperty("ignore","false").toBoolean
val datafile = NFSFileVec.make(inputfile)
val dataframe = ParseDataset.parse(Key.make("dataset-key") , datafile._key)
println("Loaded file "+inputfile+" size "+datafile.byteSize()+" Cols:"+dataframe.numCols()+" Rows:"+dataframe.numRows())
println(dataframe.anyVec().get_type_str)
for (v <- 0 to dataframe.numCols()-1) {
println(dataframe.name(v))
}
val c = dataframe.find("bad_loan")
dataframe.replace(c, dataframe.vecs()(c).toCategoricalVec())
// drop the id and member_id columns from model
dataframe.remove(dataframe.find("id"))
dataframe.remove(dataframe.find("member_id"))
// set Random Forest parameters
println("creating model parameters")
var modelparams = new DRFParameters()
var fields = modelparams.getClass.getFields
for (line <- Source.fromFile(parmsfile).getLines) {
println("Reading parameter from file: "+line)
var linedata = line.split(" ")
for(v <- fields){
if ( v.getName.matches(linedata(0))) {
val method1 = v.getDeclaringClass.getDeclaredField(linedata(0) )
method1.setAccessible(true)
println("Found "+linedata(0)+" Var "+v+" Accessable "+method1.isAccessible()+" Type "+method1.getType )
v.setAccessible(true)
v.setInt(modelparams, linedata(1).toInt)
}
}
}
// hard coded values
modelparams._train = dataframe._key
modelparams._response_column = "bad_loan"
if (ignore) {
println("Adding fields to ignore from file "+parmsfile+"FieldtoIgnore")
var ignoreNames = new Array[String](dataframe.numCols())
var in=0
for (line <- Source.fromFile(parmsfile+"FieldtoIgnore").getLines) {
ignoreNames(in) = line
in+=1
}
modelparams._ignored_columns=ignoreNames
}
println("Parameters set ")
// train model
println("Starting training")
var job: DRF = new DRF(modelparams)
var model: DRFModel = job.trainModel().get()
println("Training completed")
// training metrics
println(model._output.toString())
println("Model AUC: "+model.auc())
println(model._output._variable_importances)
// If you want to look at variables that are important and then model on them
// the following will write them out, then use only those in other model training
// handy when you have a thousand columns but want to train on only the important ones.
// Then before calling the model... call modelparams._ignored_columns= Array("inq_last_6mths")
// FileWriter
if (ignore) {
val file = new FileOutputStream(parmsfile + "FieldtoIgnore")
var n = 0
var in = 0
var ignoreNames = new Array[String](dataframe.numCols())
val fieldnames = model._output._varimp._names
println("Fields to add to _ignored_columns field")
for (i <- model._output._varimp.scaled_values()) {
if (i < 0.3) {
println(n + " = " + fieldnames(n) + " = " + i)
Console.withOut(file) {
println(fieldnames(n))
}
ignoreNames(in) = fieldnames(n)
in += 1
}
n += 1
}
println("Drop these:")
for (i <- 0 to in) {
println(fieldnames(i))
}
file.close()
println()
}
// save model
var outputfile = inputfile+"_model_pojo.txt"
var modeloutput: FileOutputStream = new FileOutputStream(outputfile)
println("Saving model to "+outputfile)
model.toJava(modeloutput, false, true)
modeloutput.close()
outputfile = inputfile+"_model_jason.txt"
modeloutput = new FileOutputStream(outputfile)
println("Saving Jason to "+outputfile)
Console.withOut(modeloutput) { println(model.toJsonString()) }
modeloutput.close()
outputfile = inputfile+"_model_mojo.zip"
modeloutput = new FileOutputStream(outputfile)
println("Saving MOJO to "+outputfile)
model.getMojo.writeTo(modeloutput)
modeloutput.close()
println(models: hex.ensemble.StackedEnsemble )
println("Completed")
H2O.shutdown(0)
}
}
Step 2: Compile and Run the MOJO¶
Open a new terminal window and change directories to the experiment folder:
$ cd experiment
Create your main program in the experiment folder by creating a new file called main.java (for example, using “vim main.java”). Include the following contents. Note that this file references the GBM model created above using R.
import java.io.*; import hex.genmodel.easy.RowData; import hex.genmodel.easy.EasyPredictModelWrapper; import hex.genmodel.easy.prediction.*; import hex.genmodel.MojoModel; public class main { public static void main(String[] args) throws Exception { EasyPredictModelWrapper model = new EasyPredictModelWrapper(MojoModel.load("GBM_model_R_1475248925871_74.zip")); RowData row = new RowData(); row.put("AGE", "68"); row.put("RACE", "2"); row.put("DCAPS", "2"); row.put("VOL", "0"); row.put("GLEASON", "6"); BinomialModelPrediction p = model.predictBinomial(row); System.out.println("Has penetrated the prostatic capsule (1=yes; 0=no): " + p.label); System.out.print("Class probabilities: "); for (int i = 0; i < p.classProbabilities.length; i++) { if (i > 0) { System.out.print(","); } System.out.print(p.classProbabilities[i]); } System.out.println(""); } }
GBM and DRF return classProbabilities, but not all MOJOs will return a classProbabilities field. Refer to the ModelPrediction definition for each algorithm to find the correct field(s) to access. This is available in the H2O-3 GitHub repo at: https://github.com/h2oai/h2o-3/tree/master/h2o-genmodel/src/main/java/hex/genmodel/easy/prediction. You can also view the hex.genmodel.easy.prediction classes in the Javadoc.
In addition to classProbabilities, you can choose to generate additional
leafNodeAssignments(GBM, DRF, Isolation Forest and XGBoost) andcontributions(GBM, DRF and XGBoost) fields. TheleafNodeAssignmentsfield will show the decision path through each tree. Thecontributionsfield will provide Shapley contributions. Note that these fields may slow down the MOJO as they add computation. Below is the Java code showing how return to both the leaf node assignment and the contributions:import java.io.*; import hex.genmodel.easy.RowData; import hex.genmodel.easy.EasyPredictModelWrapper; import hex.genmodel.easy.prediction.*; import hex.genmodel.MojoModel; public class main { public static void main(String[] args) throws Exception { EasyPredictModelWrapper.Config config = new EasyPredictModelWrapper.Config() .setModel(MojoModel.load("GBM_model_R_1475248925871_74.zip")) .setEnableLeafAssignment(true) .setEnableContributions(true); EasyPredictModelWrapper model = new EasyPredictModelWrapper(config); RowData row = new RowData(); row.put("AGE", "68"); row.put("RACE", "2"); row.put("DCAPS", "2"); row.put("VOL", "0"); row.put("GLEASON", "6"); BinomialModelPrediction p = model.predictBinomial(row); System.out.println("Has penetrated the prostatic capsule (1=yes; 0=no): " + p.label); System.out.print("Class probabilities: "); for (int i = 0; i < p.classProbabilities.length; i++) { if (i > 0) { System.out.print(","); } System.out.print(p.classProbabilities[i]); } System.out.println("Leaf node assignments: "); for (int i=0; i < p.leafNodeAssignments; i++) { if (i > 0) { System.out.print(p.leafNodeAssignments[i]); } } System.out.println(""); System.out.println("Shapley contributions: "); for (int i=0; i < p.contributions; i++) { if (i > 0) { System.out.print(","); } System.out.print(p.contributions[i]); } System.out.println(""); } }For GLRM, the returned field is the X coefficients for the archetypes by default. In addition to that, you can choose to generate the reconstructed data row as well. Again, this may slow down the MOJO due to added computation. Below is the Java code showing how to obtain both the X factors and the reconstructed data after you have generated the GLRM MOJO:
import java.io.*; import hex.genmodel.easy.RowData; import hex.genmodel.easy.EasyPredictModelWrapper; import hex.genmodel.easy.prediction.*; import hex.genmodel.MojoModel; public class main { public static void main(String[] args) throws Exception { EasyPredictModelWrapper.Config config = new EasyPredictModelWrapper.Config().setModel(MojoModel.load("GLRM_model_python_1530295749484_1.zip")).setEnableGLRMReconstrut(true); EasyPredictModelWrapper model = new EasyPredictModelWrapper(config); RowData row = new RowData(); row.put("CAPSULE", "0"); row.put("AGE", "68"); row.put("RACE", "2"); row.put("DPROS", "4"); row.put("DCAPS", "2"); row.put("PSA", "31.9"); row.put("VOL", "0"); row.put("GLEASON", "6"); DimReductionModelPrediction p = model.predictDimReduction(row); String[] colnames = model.m.getNames(); System.out.println("X coefficients for input row: "); for (int i = 0; i < p.dimensions.length; i++) { if (i > 0) { System.out.println(","); } System.out.print("Arch "+i+" coefficient: "+p.dimensions[i]); } System.out.println(""); System.out.println("Reconstructed input row: "); for (int i = 0; i < p.reconstructed.length; i++) { if (i > 0) { System.out.println(","); } System.out.print(colnames[i]+": "+p.reconstructed[i]); } System.out.println(""); }
Compile in terminal window 2.
$ javac -cp h2o-genmodel.jar -J-Xms2g -J-XX:MaxPermSize=128m main.javaRun in terminal window 2.
# Linux and OS X users $ java -cp .:h2o-genmodel.jar main # Windows users $ java -cp .;h2o-genmodel.jar main
The following output displays:
Has penetrated the prostatic capsule (1 yes; 0 no): 0 Class probabilities: 0.8059929056296662,0.19400709437033375If you have chosen to enable leaf node assignments, you will also see 100 leaf node assignments for your data row:
Has penetrated the prostatic capsule (1 yes; 0 no): 0 Class probabilities: 0.8059929056296662,0.19400709437033375 Leaf node assignments: RRRR,RRR,RRRR,RRR,RRL,RRRR,RLRR,RRR,RRR,RRR,RLRR,...For the GLRM MOJO, after running the Java code, you will see the following:
X coefficients for input row: Arch 0 coefficient: -0.5930494611027051, Arch 1 coefficient: 1.0459847877909487, Arch 2 coefficient: 0.5849220609025815 Reconstructed input row: CAPSULE: 0.5204822003860688, AGE: 10.520294102886806, RACE: 4.1422863477607645, DPROS: 2.970424071063664, DCAPS: 6.361196172145799, PSA: 1.905415090602722, VOL: 0.7123169431687857, GLEASON: 6.625024806196047
Viewing a MOJO Model¶
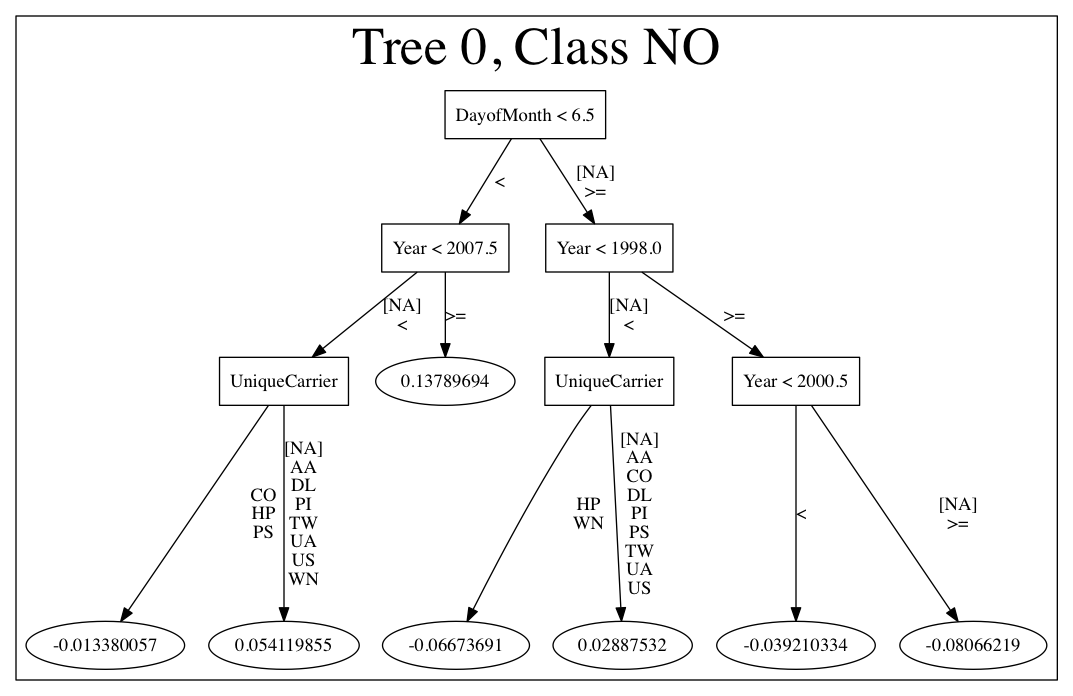
Use the PrintMojo tool to generate a graphical representation of the MOJO. PrintMojo is a java tool for converting binary MOJO files into human viewable graphs. This tool is packaged with H2O and produces an output that “dot” (which is part of Graphviz) can turn into an image. (See the Graphviz home page for more information.)
Here is an example output for a GBM model:
The following options can be specified with PrintMojo:
--input(or-i): Required. Sepcify the MOJO file name--output(or-o): Optionally specify the output file name. This is taken as a directory name in the case of .png format and multiple trees to visualize. This defaults to stdout.--format: Optionally specify the output format. Available formats include dot (default), json, raw, and png. Note that for .png output, Java 8 is the minimum Java requirement.--tree: Optionally specify the tree number to print. This defaults to “all”.--levels: Optionaly specify the number of levels per edge to print. This defaults to 10.--title: Optionally force the title of the tree graph.--detail: Specifies to print additional information such as node numbers.--decimalplaces(or-d): Allows you to control the number of decimal points shown for numbers.--fontsize(or-f): Controls the font size. The default font size is 14. When using this option, be careful not to choose a font size that is so large that you cannot see your whole tree. We recommend using a font size no larger than 20.--internal: Optional. Internal H2O representation of the decision tree (splits etc.). This is used for generating the Graphviz format.
The following code snippet shows how to download a MOJO and run the PrintMojo tool from the command line to make a .png file. Note that this requires that Graphviz is installed.
library(h2o)
h2o.init()
df <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
model <- h2o.gbm(model_id = "model",
training_frame = df,
x = c("Year", "Month", "DayofMonth", "DayOfWeek", "UniqueCarrier"),
y = "IsDepDelayed",
max_depth = 3,
ntrees = 5)
h2o.download_mojo(model, getwd(), FALSE)
# In another terminal window, download and extract the
# latest stable h2o.jar from http://www.h2o.ai/download/
cd ~/Downloads
unzip h2o-3.30.0.5.zip
cd h2o-3.30.0.5
# Run the PrintMojo tool from the command line.
# This requires that graphviz is installed.
brew install graphviz # if not already installed
java -cp h2o.jar hex.genmodel.tools.PrintMojo --tree 0 -i "path/to/model.zip" -o model.gv -f 20 -d 3
dot -Tpng model.gv -o model.png
open model.png
import h2o
h2o.init()
from h2o.estimators.gbm import H2OGradientBoostingEstimator
df = h2o.import_file("http://s3.amazonaws.com/h2o-public-test-data/smalldata/airlines/allyears2k_headers.zip")
predictors = ["Year", "Month", "DayofMonth", "DayOfWeek", "UniqueCarrier"]
response = "IsDepDelayed"
model = H2OGradientBoostingEstimator(max_depth=3, ntrees=5)
model.train(x = predictors,
y = response,
training_frame = df)
model.download_mojo(path="/path/to/mojo/zip", get_genmodel_jar=False, genmodel_name=genmodel_path)
# In another terminal window, download and extract the
# latest stable h2o.jar from http://www.h2o.ai/download/
cd ~/Downloads
unzip h2o-3.30.0.5.zip
cd h2o-3.30.0.5
# Run the PrintMojo tool from the command line.
# This requires that graphviz is installed.
brew install graphviz # example for Mac OsX if not already installed
java -cp h2o.jar hex.genmodel.tools.PrintMojo --tree 0 -i "path/to/model.zip" -o model.gv -f 20 -d 3
dot -Tpng model.gv -o model.png
open model.png
If you cannot install Graphviz on your environment, another option is to produce a picture output directly with PrintMojo. This option requires Java 8 or higher and uses the h2o-genmodel.jar file.
Extract h2o-genmodel.jar from the running h2o instance:
# In one terminal window run: java -jar h2o.jar # While still running H2O in the first terminal window, # in the second terminal window run: curl http://localhost:54321/3/h2o-genmodel.jar > h2o-genmodel.jar
Run the PrintMojo tool on the command line to make a .png file without using Graphviz.
# Download the latest stable h2o release from http://www.h2o.ai/download/ # and run the PrintMojo tool from the command line. java -cp h2o-genmodel.jar hex.genmodel.tools.PrintMojo --tree 0 -i "/path/to/model.zip" -o tree.png --format png open tree.png
Printing GBM MOJOs¶
The print_mojo function allows a model to be converted to machine readable representation - either to JSON for further processing or dot for rendering images.
When running print_mojo, the following can be specified:
mojo_path: The path to the MOJO archive on the user’s local filesystem. This defaults to the user’s home directory.format: Specify an output format of eitherjson(default) ordot.tree_index: Specify the index of the tree to print. This can only be used withdotformat.
The output is a string respresentation of the MOJO in either JSON or dot format.
Note: print_mojo can only be used with the Python client. It is not supported in R.
Example¶
import h2o
import json
from h2o.estimators import H2OGradientBoostingEstimator
h2o.init()
# Import the prostate dataset
prostate = h2o.import_file("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv")
# Define the factor and parameters
prostate["CAPSULE"] = prostate["CAPSULE"].asfactor()
ntrees = 20
learning_rate = 0.1
depth = 5
min_rows = 10
# Build & train the GBM model
gbm_h2o = H2OGradientBoostingEstimator(ntrees = ntrees,
learn_rate = learning_rate,
max_depth = depth,
min_rows = min_rows,
distribution = "bernoulli")
gbm_h2o.train(x = list(range(1,prostate.ncol)), y = "CAPSULE", training_frame = prostate)
# Print all
mojo_path = gbm_h2o.download_mojo()
mojo_str = h2o.print_mojo(mojo_path)
mojo_dict = json.loads(mojo_str)
FAQ¶
Are MOJOs thread safe?
Yes, all of H2O-3 MOJOs are thread safe.
How can I use an XGBoost MOJO with Maven?
If you declare a dependency on h2o-genmodel, then you also have to include the h2o-genmodel-ext-xgboost dependency if you are planning to use XGBoost models. For example:
<groupId>ai.h2o</groupId> <artifactId>xgboost-mojo-example</artifactId> <version>1.0-SNAPSHOT</version> dependency> <groupId>ai.h2o</groupId> <artifactId>h2o-genmodel-ext-xgboost</artifactId> <version>3.30.0.5</version> </dependency> <dependency> <groupId>ai.h2o</groupId> <artifactId>h2o-genmodel</artifactId> <version>3.30.0.5</version> </dependency>
POJO Quick Start¶
This section describes how to build and implement a POJO to use predictive scoring. Java developers should refer to the Javadoc for more information, including packages.
Notes:
POJOs are not supported for source files larger than 1G. For more information, refer to the POJO FAQ section below. POJOs are also not supported for GLRM or Stacked Ensembles models.
POJO predict cannot parse columns enclosed in double quotes (for example, “”2””).
What is a POJO?¶
H2O allows you to convert the models you have built to a Plain Old Java Object (POJO), which can then be easily deployed within your Java app and scheduled to run on a specified dataset.
POJOs allow users to build a model using H2O and then deploy the model to score in real-time, using the POJO model or a REST API call to a scoring server.
The only compilation and runtime dependency for a generated model is the h2o-genmodel.jar file produced as the build output of these packages. This file is a library that supports scoring, and it contains the base classes from which the POJO is derived from. (You can see “extends GenModel” in a pojo class. The GenModel class is part of this library.) The h2o-genmodel.jar file is required when POJO models are deployed to production.
Building a POJO¶
The example code below shows how to start H2O Flow, compile and run an example Flow, and then compile and run the POJO. This example uses GBM, but any supported algorithm can be used to build a model and run the POJO.
Start H2O in terminal window #1:
$ java -jar h2o.jarBuild a model using your web browser:
Go to http://localhost:54321
Click View Example Flows near the right edge of the screen. Here is a screenshot of what to look for:
Click
GBM_Airlines_Classification.flowIf a confirmation prompt appears asking you to “Load Notebook”, click it.
From the “Flow” menu choose the “Run all cells” option
Scroll down and find the “Model” cell in the notebook. Click on the Download POJO button as shown in the following screenshot:
Note: The instructions below assume that the POJO model was downloaded to the “Downloads” folder.
Download model pieces in a new terminal window. Note that H2O must still be running in terminal window #1:
$ mkdir experiment $ cd experiment $ mv ~/Downloads/gbm_pojo_test.java . $ curl http://localhost:54321/3/h2o-genmodel.jar > h2o-genmodel.jar
Create your main program in terminal window #2 by creating a new file called main.java (
vim main.java) with the following contents:import java.io.*; import hex.genmodel.easy.RowData; import hex.genmodel.easy.EasyPredictModelWrapper; import hex.genmodel.easy.prediction.*; public class main { private static String modelClassName = "gbm_pojo_test"; public static void main(String[] args) throws Exception { hex.genmodel.GenModel rawModel; rawModel = (hex.genmodel.GenModel) Class.forName(modelClassName).newInstance(); EasyPredictModelWrapper model = new EasyPredictModelWrapper(rawModel); RowData row = new RowData(); row.put("Year", "1987"); row.put("Month", "10"); row.put("DayofMonth", "14"); row.put("DayOfWeek", "3"); row.put("CRSDepTime", "730"); row.put("UniqueCarrier", "PS"); row.put("Origin", "SAN"); row.put("Dest", "SFO"); BinomialModelPrediction p = model.predictBinomial(row); System.out.println("Label (aka prediction) is flight departure delayed: " + p.label); System.out.print("Class probabilities: "); for (int i = 0; i < p.classProbabilities.length; i++) { if (i > 0) { System.out.print(","); } System.out.print(p.classProbabilities[i]); } System.out.println(""); } }
Compile the POJO in terminal window 2:
$ javac -cp h2o-genmodel.jar -J-Xmx2g -J-XX:MaxPermSize=128m gbm_pojo_test.java main.java
Run the POJO in terminal window 2.
For Linux and OS X users:
$ java -cp .:h2o-genmodel.jar mainFor Windows users:
$ java -cp .;h2o-genmodel.jar mainThe following output displays:
Label (aka prediction) is flight departure delayed: YES Class probabilities: 0.4319916897116479,0.5680083102883521
Extracting Models from H2O¶
Generated models can be extracted from H2O in the following ways:
From the H2O Flow Web UI¶
When viewing a model, click the Download POJO button at the top of the model cell, as shown in the example in the Quick start section. You can also preview the POJO inside Flow, but it will only show the first thousand lines or so in the web browser, truncating large models.
From R or Python¶
The following code snippets show an example of H2O building a model and downloading its corresponding POJO from an R script and a Python script.
library(h2o)
h2o.init()
path <- system.file("extdata", "prostate.csv", package = "h2o")
h2o_df <- h2o.importFile(path)
h2o_df$CAPSULE <- as.factor(h2o_df$CAPSULE)
model <- h2o.glm(y = "CAPSULE",
x = c("AGE", "RACE", "PSA", "GLEASON"),
training_frame = h2o_df,
family = "binomial")
h2o.download_pojo(model)
import h2o
h2o.init()
from h2o.estimators.glm import H2OGeneralizedLinearEstimator
path = "http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip"
h2o_df = h2o.import_file(path)
h2o_df['CAPSULE'] = h2o_df['CAPSULE'].asfactor()
model = H2OGeneralizedLinearEstimator(family = "binomial")
model.train(y = "CAPSULE",
x = ["AGE", "RACE", "PSA", "GLEASON"],
training_frame = h2o_df)
h2o.download_pojo(model)
Use Cases¶
The following use cases are demonstrated with code examples:
Reading new data from a CSV file and predicting on it: The PredictCsv class is used by the H2O test harness to make predictions on new data points.
Getting a new observation from a JSON request and returning a prediction
Calling a user-defined function directly from hive: See the H2O-3 training github repository.
FAQ¶
Are POJOs thread safe?
Yes, all of H2O-3 POJOs are thread safe.
How do I score new cases in real-time in a production environment?
If you’re using the UI, click the Preview POJO button for your model. This produces a Java class with methods that you can reference and use in your production app.
What kind of technology would I need to use?
Anything that runs in a JVM. The POJO is a standalone Java class with no dependencies on H2O.
How should I format my data before calling the POJO?
Here are our requirements (assuming you are using the “easy” Prediction API for the POJO as described in the Javadoc).
Input columns must only contain categorical levels that were seen during training
Any additional input columns not used for training are ignored
If no input column is specified, it will be treated as an
NASome models do not handle NAs well (e.g., GLM)
Any transformations applied to data before model training must also be applied before calling the POJO predict method
How do I run a POJO on a Spark Cluster?
The POJO provides just the math logic to do predictions, so you won’t find any Spark (or even H2O) specific code there. If you want to use the POJO to make predictions on a dataset in Spark, create a map to call the POJO for each row and save the result to a new column, row-by-row.
How do I communicate with a remote cluster using the REST API?
You can dl the POJO using the REST API but when calling the POJO predict function, it’s in the same JVM, not across a REST API.
Is it possible to make predictions using my H2O cluster with the REST API?
Yes, but this way of making predictions is separate from the POJO. For more information about in-H2O predictions (as opposed to POJO predictions), see the documentation for the H2O REST API endpoint /3/Predictions.
Why did I receive the following error when trying to compile the POJO?
The following error is generated when the source file is larger than 1G.
Michals-MBP:b michal$ javac -cp h2o-genmodel.jar -J-Xmx2g -J-XX:MaxPermSize=128m drf_b9b9d3be_cf5a_464a_b518_90701549c12a.java An exception has occurred in the compiler (1.7.0_60). Please file a bug at the Java Developer Connection (http://java.sun.com/webapps/bugreport) after checking the Bug Parade for duplicates. Include your program and the following diagnostic in your report. Thank you. java.lang.IllegalArgumentException at java.nio.ByteBuffer.allocate(ByteBuffer.java:330) at com.sun.tools.javac.util.BaseFileManager$ByteBufferCache.get(BaseFileManager.java:308) at com.sun.tools.javac.util.BaseFileManager.makeByteBuffer(BaseFileManager.java:280) at com.sun.tools.javac.file.RegularFileObject.getCharContent(RegularFileObject.java:112) at com.sun.tools.javac.file.RegularFileObject.getCharContent(RegularFileObject.java:52) at com.sun.tools.javac.main.JavaCompiler.readSource(JavaCompiler.java:571) at com.sun.tools.javac.main.JavaCompiler.parse(JavaCompiler.java:632) at com.sun.tools.javac.main.JavaCompiler.parseFiles(JavaCompiler.java:909) at com.sun.tools.javac.main.JavaCompiler.compile(JavaCompiler.java:824) at com.sun.tools.javac.main.Main.compile(Main.java:439) at com.sun.tools.javac.main.Main.compile(Main.java:353) at com.sun.tools.javac.main.Main.compile(Main.java:342) at com.sun.tools.javac.main.Main.compile(Main.java:333) at com.sun.tools.javac.Main.compile(Main.java:76) at com.sun.tools.javac.Main.main(Main.java:61)
Example Design Patterns¶
Here is a collection of example design patterns for how to productionize H2O.
Consumer loan application¶
Characteristic |
Value |
|---|---|
Pattern name |
Jetty servlet |
Example training language |
R |
Example training data source |
CSV file |
Example scoring data source |
User input to Javascript application running in browser |
Scoring environment |
REST API service provided by Jetty servlet |
Scoring engine |
H2O POJO |
Scoring latency SLA |
Real-time |
Craigslist application¶
Characteristic |
Value |
|---|---|
Pattern name |
Sparkling water streaming |
Example training language |
Scala |
Example training data source |
CSV file |
Example scoring data source |
User input to Javascript application running in browser |
Scoring engine |
H2O cluster |
Scoring latency SLA |
Real-time |
Resource |
Location |
|---|---|
Git repos |
|
Blogs |
|
Slides |
http://www.slideshare.net/0xdata/sparkling-water-ask-craig http://www.slideshare.net/0xdata/sparkling-water-applications-meetup-072115 |
Malicious domain application¶
Characteristic |
Value |
|---|---|
Pattern name |
AWS Lambda |
Example training language |
Python |
Example training data source |
CSV file |
Example scoring data source |
User input to Javascript application running in browser |
Scoring environment |
AWS Lambda REST API endpoint |
Scoring engine |
H2O POJO |
Scoring latency SLA |
Real-time |
Storm bolt¶
Characteristic |
Value |
|---|---|
Pattern name |
Storm bolt |
Example training language |
R |
Example training data source |
CSV file |
Example scoring data source |
Storm spout |
Scoring environment |
POJO embedded in a Storm bolt |
Scoring engine |
H2O POJO |
Scoring latency SLA |
Real-time |
Resource |
Location |
|---|---|
Git repos |
https://github.com/h2oai/h2o-tutorials/tree/master/tutorials/streaming/storm |
Tutorials |
http://docs.h2o.ai/h2o-tutorials/latest-stable/tutorials/streaming/storm/index.html |
Invoking POJO directly in R¶
Characteristic |
Value |
|---|---|
Pattern name |
POJO in R |
Example training language |
R |
Example training data source |
(Need example) |
Example scoring data source |
(Need example) |
Scoring environment |
R |
Scoring engine |
H2O POJO |
Scoring latency SLA |
Batch |
Hive UDF¶
Characteristic |
Value |
|---|---|
Pattern name |
Hive UDF |
Example training language |
R |
Example training data source |
HDFS directory with hive part files output by a SELECT |
Example scoring data source |
Hive |
Scoring environment |
Hive SELECT query (parallel MapReduce) running UDF |
Scoring engine |
H2O POJO |
Scoring latency SLA |
Batch |
Resource |
Location |
|---|---|
Git repos |
https://github.com/h2oai/h2o-tutorials/tree/master/tutorials/hive_udf_template |
POJO Tutorial |
|
MOJO Tutorial |
MOJO as a JAR Resource¶
Characteristic |
Value |
|---|---|
Pattern name |
MOJO JAR |
Example training language |
R |
Example training data source |
Iris |
Example scoring data source |
Single Row |
Scoring environment |
Portable |
Scoring engine |
H2O MOJO |
Scoring latency SLA |
Real-time example, but can be adapted (use in Hive UDF etc.) |
Resource |
Location |
|---|---|
Git repos |
https://github.com/h2oai/h2o-tutorials/tree/master/tutorials/mojo-resource |
Steam Scoring Server from H2O.ai¶
Characteristic |
Value |
|---|---|
Pattern name |
Steam |
Scoring data source |
REST API client |
Scoring environment |
Steam scoring server |
Scoring engine |
H2O POJO |
Scoring latency SLA |
Real-time |
Resource |
Location |
|---|---|
Web sites |
Scoring Server on AWS¶
You can deploy a RESTful server on AWS using the marketplace AMI (H2O Inference server - Hourly). Notice that this is a paid AMI.
Transfer the MOJO file into the /tmp folder of this instance before launching. If your MOJO is in S3, assign a role that provides S3 access to the instance.
Run following bash script as “userdata” to transfer your MOJO into the instance before you launch the instance. Be sure you change the mojofile path below.
#cloud-boothook
#!/bin/bash
export mojofile="s3://yourbucket/yourmojo.zip"
aws s3 cp $mojofile /tmp/pipeline.mojo
After this instance has launched, you can make real time inference using the following command. Remember to change the IP address. Input data is provided through the row parameter in the URL.
curl "http://<yourIP>:8080/model?type=1&row=2000,2000"