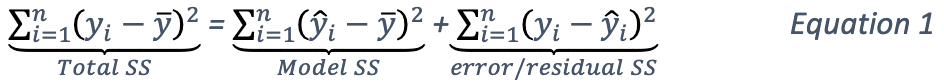ANOVA GLM¶
H2O ANOVA GLM is used to calculate Type III SS (sum of squares) which is used to investigate the contributions of individual predictors and their interactions to a model. Predictors or interactions with negligible contributions to the model will have high p-values while those with more contributions will have low p-values. We use predictors to express individual predictors or interactions of predictors.
Since ANOVA GLM is mainly used to investigate the contribution of each predictor or interaction, scoring, MOJO, and cross-validation are not supported.
Defining an ANOVA GLM Model¶
ANOVA GLM uses a similar set of parameters to GLM.
model_id: (Optional) Specify a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Specify the dataset used to build the model. NOTE: In Flow, if you click the Build a model button from the
Parsecell, the training frame is entered automatically.y: (Required) Specify the column to use as the dependent variable. The data can be numeric or categorical.
x: Specify a vector containing the names or indices of the predictor variables to use when building the model. If
xis missing, then all columns exceptyare used.ignored_columns: (Optional, Python and Flow only) Specify the column or columns to be excluded from the model. In Flow, click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Specify whether to ignore constant training columns, since no information can be gained from them. This option is defaults to true (enabled).
seed: Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations. This value defaults to -1 (time-based random number).
score_each_iteration: (Optional) Specify whether to score during each iteration of the model training. This value is disabled by default.
offset_column: Specify a column to use as the offset.
weights_column: Specify a column to use for the observation weights, which are used for bias correction. The specified
weights_columnmust be included in the specifiedtraining_frame.Python only: To use a weights column when passing an H2OFrame to
xinstead of a list of column names, the specifiedtraining_framemust contain the specifiedweights_column.Note: Weights are per-row observation weights and do not increase the size of the data frame. This is typically the number of times a row is repeated, but non-integer values are supported as well. During training, rows with higher weights matter more, due to the larger loss function pre-factor.
balance_classes: Specify whether to oversample the minority classes to balance the class distribution. This option is defaults to false (not enabled), and can increase the data frame size. This option is only applicable for classification. Majority classes can be undersampled to satisfy the max_after_balance_size parameter.
stopping_rounds: Stops training when the option selected for stopping_metric doesn’t improve for the specified number of training rounds, based on a simple moving average. This option is defaults 0 (no early stopping). The metric is computed on the validation data (if provided); otherwise, training data is used.
stopping_metric: Specify the metric to use for early stopping. The available options are:
AUTO: This defaults tologlossfor classification,deviancefor regression, andanomaly_scorefor Isolation Forest.anomaly_score(Isolation Forest only)devianceloglossMSERMSEMAERMSLEAUC(area under the ROC curve)AUCPR(area under the Precision-Recall curve)lift_top_groupmisclassificationmean_per_class_errorcustom(Python client only)custom_increasing(Python client only)
stopping_tolerance: Specify the relative tolerance for the metric-based stopping to stop training if the improvement is less than this value. This value defaults to 0.001.
early_stopping: Specify whether to stop early when there is no more relative improvement on the training or validation set. This option is defaults to False (disabled).
max_runtime_secs: Maximum allowed runtime in seconds for model training. This defaults to 0 (unlimited).
class_sampling_factors: Specify the per-class (in lexicographical order) over/under-sampling ratios. By default, these ratios are automatically computed during training to obtain the class balance. Note that this requires
balance_classes=true.max_after_balance_size: Specify the maximum relative size of the training data after balancing class counts (balance_classes must be enabled). The value can be less than 1.0 and defaults to 5.0.
family: Specify the model type. Use binomial for classification with logistic regression, others are for regression problems. One of
"auto"``(default), ``"gaussian","binomial",fractionalbinomial,"quasibinomial","poisson","gamma","tweedie","negativebinomial".tweedie_variance_power: (Only applicable if Tweedie is specified for Family) Specify the Tweedie variance power (defaults to 0).
tweedie_link_power: (Only applicable if Tweedie is specified for Family) Specify the Tweedie link power (defaults to 1).
theta: Theta value (equal to 1/r) for use with the negative binomial family. This value must be > 0 and defaults to 0.
solver: Specify the solver to use (
AUTO,IRLSM(default),L_BFGS,COORDINATE_DESCENT_NAIVE,COORDINATE_DESCENT,GRADIENT_DESCENT_LH, orGRADIENT_DESCENT_SQERR). IRLSM is fast on problems with a small number of predictors and for lambda search with L1 penalty, while L_BFGS scales better for datasets with many columns. COORDINATE_DESCENT is IRLSM with the covariance updates version of cyclical coordinate descent in the innermost loop. COORDINATE_DESCENT_NAIVE is IRLSM with the naive updates version of cyclical coordinate descent in the innermost loop. GRADIENT_DESCENT_LH and GRADIENT_DESCENT_SQERR can only be used with the Ordinal family. AUTO will set the solver based on the given data and other parameters.alpha: Specify the regularization distribution between L1 and L2. The default value of alpha is 0 when
solver = 'L-BFGS', overwise it is 0.5.lambda: Specify the regularization strength. Defaults to
[0.0].lambda_search: Specify whether to enable lambda search, starting with lambda max (the smallest \(\lambda\) that drives all coefficients to zero). If you also specify a value for
lambda_min_ratio, then this value is interpreted as lambda min. If you do not specify a value forlambda_min_ratio, then GAM will calculate the minimum lambda. This option is defaults to false (not enabled).standardize: Specify whether to standardize the numeric columns to have a mean of zero and unit variance. This option defaults to True.
missing_values_handling: Specify how to handle missing values (Skip, MeanImputation, or PlugValues). This value defaults to MeanImputation.
plug_values: When
missing_values_handling="PlugValues", specify a single row frame containing values that will be used to impute missing values of the training/validation frame.compute_p_values: Request computation of p-values. P-values only work with the
IRLSMsolver and no regularization. Defaults to True.non_negative: Specify whether to force coefficients to have non-negative values. This option is defaults to false.
max_iterations: Specify the number of training iterations (defaults to 0).
link: Specify a link function (
Identity,Family_Default,Logit,Log,Inverse,Tweedie, orOlogit). The default value is Family_Default.prior: Specify prior probability for p(y==1). Use this parameter for logistic regression if the data has been sampled and the mean of response does not reflect reality. This value must be a value in the range (0,1) or set to -1 (disabled). This option is set to 0 by default.
Note: This is a simple method affecting only the intercept. You may want to use weights and offset for a better fit.
type: Refer to the SS type 1, 2, 3, or 4.
Note: We are currently only supporting 3.
highest_interaction_term: This limits the number of interaction terms (i.e. 2 means interaction between 2 columns only, 3 for three columns, etc.). Defaults to 2.
save_transformed_framekeys: Set to True to save the keys of transformed predictors and interaction column. Defaults to False.
To demonstrate what Type III SS is and how it is implemented, here is an example of regression with two categorical predictors:
note: This algorithm will support multiple categorical/numerical columns and other families as well; we just need to replace the SS with the residual deviance for other families.
SS (Sum of Squares)¶
In Analysis of Variance (ANOVA), the partition of the response variable sum of squares in a linear model is described as “explained” and “unexplained” components. Consider a dataset generated by
\[y_i = x^T_i\beta + \epsilon_i\]
where
\(y_i\) is the response variable;
\(x^T_i = [1,x_{i1},...,x_{im}]\) are the predictors;
\(\beta = [\beta_0, \beta_1,..., \beta_m]\) are the system parameters;
\(\epsilon_i {\text{ ~ }} N(0,\sigma^2)\).
The total sum of squares of this dataset can be decomposed as follows:
where
\(\bar{y} = {\frac{1}{n}}{\sum^n_{i=1}}y_i\);
\(\hat{y_i} = x^T_i \hat{\beta} {\text{ and }} \hat{\beta} = (X^TX)^{-1}X^TY, X = {\begin{bmatrix}1^T \\ x^T_1 \\ x^T_2 \\ ... \\ X^T_m\end{bmatrix}}, Y = {\begin{bmatrix}y_1 \\ ... \\ y_n\end{bmatrix}}\).
Generally, addition of a new predictor to a model will increase the model SS and reduce the error or residual SS.
The model SS by itself is not useful. However, if you have multiple models, the difference in model SS between two models can be used to determine model performance gain/loss.
Type III SS Calculation¶
The Type III SS calculation can be illustrated using two predictors (C,R). Let
\(SS(C,R,C:R)\) denote the model sum of squares for GLM with predictors C,R and the interaction of C and R;
\(SS(C,R)\) denote the model sum of squares for GLM with predictors C,R only;
\(SS(R,C:R)\) denote the model sum of squares for GLM with predictors R and the interaction of C and R;
\(SS(C,C:R)\) denote the model sum of squares for GLM with predictors C and the interaction of C and R.
Type III SS calculation refers to the incremental sum of squares by taking the difference between the model sum of squares for alternative models:
\(SS(C|R,C:R) = SS(C,R,C:R) - SS(R,C:R) = error SS(R,C:R) - error SS(C,R,C:R)\);
\(SS(R|C,C:R) = SS(C,R,C:R) - SS(C,C:R) = error SS(C,C:R) - error SS(C,R,C:R)\);
\(SS(C:R|R,C) = SS(C,R,C:R) - SS(R,C) = error SS(R,C) - error SS(C,R,C:R)\).
The second part of the equations can be derived from Equation 1. Note that the \(error SS\) is just the residual deviance of the models.
The same procedure applies if there are more predictors. In general, to calculate the Type III SS, we build the model with all the predictors and all the predictor interactions and compare the full model to taking out either one predictor or one interaction. For example, if there are three predictors (R,C,S), then all of the following predictors can be found in the model: R, C, S, R:C, R:S, C:S, R:C:S. Hence, we calculate the difference in SS of the full model with one predictor out of the seven predictors left out. In addition, to control the number of predictors in the interaction, the parameter highest_interaction_term is added to limit the number of predictors involved in an interaction. Using the example of three predictors, if highest_interaction_term=2, the predictors used in building the full model will only be R, C, S, R:C, R:S, C:S. The interaction term R:C:S will be excluded for it has 3 predictors which is not allowed in this case.
The calculation of the SS difference is then used to estimate how important the predictor that is left out is. To do this, F-tests are used. Using the example of two categorical predictors R with r levels, C with c levels, the following table will be generated for a dataset of n rows:
Source |
Degree of freedom |
Model SS |
Hypothesis |
F |
|---|---|---|---|---|
R |
\(r-1\) |
\(SS(R|C,R:C)\) |
Coefficients for R are zero. |
\({\frac{SS(R|C,R:C)(n-r*C)}{(r-1)*errorSS}}\) |
C |
\(c-1\) |
\(SS(C|R,R:C)\) |
Coefficients for C are zero. |
\({\frac{SS(C|R,R:C)(n-r*c)}{(c-1)*errorSS}}\) |
R:C Interaction |
\((r-1)*(c-1)\) |
\(SS(R:C|R,C)\) |
Coefficients for interaction R:C are zero. |
\({\frac{SS(R:C|R,C)(n-r*c)}{(r-1)(c-1)*errorSS}}\) |
Residual SS |
\(n-r*c\) |
\(errorSS\) of full model |
||
Total: |
\(n-1\) |
Finally, to answer the question that certain coefficients should be zero, we calculate the p-value from the F-tests just like the p-value calculation with a Gaussian distribution. In this case, we assume that the distribution of F is the F statistic. If the p-value calculated is small, you reject the hypothesis that the set of parameters associated with a predictor should be set to zero.
Examples¶
library(h2o) h2o.init() # Import the prostate dataset: train <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate_complete.csv.zip") # Set the predictors and response: x <- c("AGE", "VOL", "DCAPS") y <- "CAPSULE" # Build and train the model: anova_model <- h2o.anovaglm(y = 'CAPSULE', x = c('AGE','VOL','DCAPS'), training_frame = train, family = "binomial", missing_values_handling="MeanImputation") # Check the model summary: summary(anova_model)import h2o h2o.init() from h2o.estimators import H2OANOVAGLMEstimator #Import the prostate dataset train = h2o.import_file("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate_complete.csv.zip") # Set the predictors and response: x = ['AGE','VOL','DCAPS'] y = 'CAPSULE' # Build and train the model: anova_model = H2OANOVAGLMEstimator(family='binomial', lambda_=0, missing_values_handling="skip") anova_model.train(x=x, y=y, training_frame=train) # Get the model summary: anova_model.summary()