Welcome to H2O 3.0
- New Users
- Experienced Users
- Enterprise Users
- Sparkling Water Users
- Python Users
- R Users
- API Users
- Java Users
- Developers
Welcome to the H2O documentation site! Depending on your area of interest, select a learning path from the links above.
We’re glad you’re interested in learning more about H2O - if you have any questions or need general support, please email them to our Google Group, h2ostream or post them on our Google groups forum, h2ostream. This is a public forum, so your question will be visible to other users.
Note: To join our Google group on h2ostream, you need a Google account (such as Gmail or Google+). On the h2ostream page, click the Join group button, then click the New Topic button to post a new message. You don’t need to request or leave a message to join - you should be added to the group automatically.
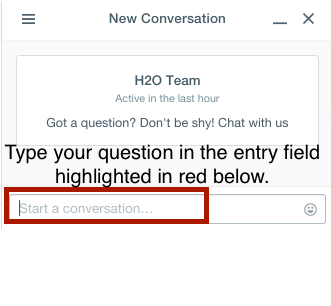
We welcome your feedback! Please let us know if you have any questions or comments about H2O by clicking the chat balloon button in the lower-right corner in Flow (H2O’s web UI).
Type your question in the entry field that appears at the bottom of the sidebar and you will be connected with an H2O expert who will respond to your query in real time.
New Users
If you’re just getting started with H2O, here are some links to help you learn more:
Recommended Systems: This one-page PDF provides a basic overview of the operating systems, languages and APIs, Hadoop resource manager versions, cloud computing environments, browsers, and other resources recommended to run H2O. At a minimum, we recommend the following for compatibility with H2O:
- Operating Systems: Windows 7 or later; OS X 10.9 or later, Ubuntu 12.04, or RHEL/CentOS 6 or later
- Languages: Java 7 or later; Scala v 2.10 or later; R v.3 or later; Python 2.7.x or 3.5.x (Scala, R, and Python are not required to use H2O unless you want to use H2O in those environments, but Java is always required)
- Browsers: Latest version of Chrome, Firefox, Safari, or Internet Explorer (An internet browser is required to use H2O’s web UI, Flow)
- Hadoop: Cloudera CDH 5.2 or later (5.3 is recommended); MapR v.3.1.1 or later; Hortonworks HDP 2.1 or later (Hadoop is not required to run H2O unless you want to deploy H2O on a Hadoop cluster)
- Spark: v 1.3 or later (Spark is only required if you want to run Sparkling Water)
Downloads page: First things first - download a copy of H2O here by selecting a build under “Download H2O” (the “Bleeding Edge” build contains the latest changes, while the latest alpha release is a more stable build), then use the installation instruction tabs to install H2O on your client of choice (standalone, R, Python, Hadoop, or Maven) .
For first-time users, we recommend downloading the latest alpha release and the default standalone option (the first tab) as the installation method. Make sure to install Java if it is not already installed.
Tutorials: To see a step-by-step example of our algorithms in action, select a model type from the following list:
Getting Started with Flow: This document describes our new intuitive web interface, Flow. This interface is similar to IPython notebooks, and allows you to create a visual workflow to share with others.
Launch from the command line: This document describes some of the additional options that you can configure when launching H2O (for example, to specify a different directory for saved Flow data, allocate more memory, or use a flatfile for quick configuration of a cluster).
Algorithms: This document describes the science behind our algorithms and provides a detailed, per-algo view of each model type.
Experienced Users
If you’ve used previous versions of H2O, the following links will help guide you through the process of upgrading to H2O 3.0.
Recommended Systems: This one-page PDF provides a basic overview of the operating systems, languages and APIs, Hadoop resource manager versions, cloud computing environments, browsers, and other resources recommended to run H2O.
Migration Guide: This document provides a comprehensive guide to assist users in upgrading to H2O 3.0. It gives an overview of the changes to the algorithms and the web UI introduced in this version and describes the benefits of upgrading for users of R, APIs, and Java.
Porting R Scripts: This document is designed to assist users who have created R scripts using previous versions of H2O. Due to the many improvements in R, scripts created using previous versions of H2O need some revision to work with H2O 3.0. This document provides a side-by-side comparison of the changes in R for each algorithm, as well as overall structural enhancements R users should be aware of, and provides a link to a tool that assists users in upgrading their scripts.
Recent Changes: This document describes the most recent changes in the latest build of H2O. It lists new features, enhancements (including changed parameter default values), and bug fixes for each release, organized by sub-categories such as Python, R, and Web UI.
H2O Classic vs H2O 3.0: This document presents a side-by-side comparison of H2O 3.0 and the previous version of H2O. It compares and contrasts the features, capabilities, and supported algorithms between the versions. If you’d like to learn more about the benefits of upgrading, this is a great source of information.
Algorithms Roadmap: This document outlines our currently implemented features and describes which features are planned for future software versions. If you’d like to know what’s up next for H2O, this is the place to go.
Contributing code: If you’re interested in contributing code to H2O, we appreciate your assistance! This document describes how to access our list of Jiras that are suggested tasks for contributors and how to contact us.
Enterprise Users
If you’re considering using H2O in an enterprise environment, you’ll be happy to know that the H2O platform is supported on all major Hadoop distributions including Cloudera Enterprise, Hortonworks Data Platform and the MapR Apache Hadoop Distribution.
H2O can be deployed in-memory directly on top of existing Hadoop clusters without the need for data transfers, allowing for unmatched speed and ease of use. To ensure the integrity of data stored in Hadoop clusters, the H2O platform supports native integration of the Kerberos protocol.
For additional sales or marketing assistance, please email sales@h2o.ai.
Recommended Systems: This one-page PDF provides a basic overview of the operating systems, languages and APIs, Hadoop resource manager versions, cloud computing environments, browsers, and other resources recommended to run H2O.
H2O Enterprise Edition: This web page describes the benefits of H2O Enterprise Edition.
Security: This document describes how to use the security features (available only in H2O Enterprise Edition).
- How to Pass S3 Credentials to H2O: This document describes the necessary step of passing your S3 credentials to H2O so that H2O can be used with AWS, as well as how to run H2O on an EC2 cluster.
Click here to view instructions on how to set up H2O using Hadoop.
Running H2O on Hadoop: This document describes how to run H2O on Hadoop.
Sparkling Water Users
- Getting Started with Sparkling Water
- Sparkling Water Blog Posts
- Sparkling Water Meetup Slide Decks
- PySparkling
Sparkling Water is a gradle project with the following submodules:
- Core: Implementation of H2OContext, H2ORDD, and all technical integration code
- Examples: Application, demos, examples
- ML: Implementation of MLLib pipelines for H2O algorithms
- Assembly: Creates “fatJar” composed of all other modules
- py: Implementation of (h2o) Python binding to Sparkling Water
The best way to get started is to modify the core module or create a new module, which extends a project.
Users of our Spark-compatible solution, Sparkling Water, should be aware that Sparkling Water is only supported with the latest version of H2O. For more information about Sparkling Water, refer to the following links.
Sparkling Water is versioned according to the Spark versioning, so make sure to use the Sparkling Water version that corresponds to the installed version of Spark:
- Use Sparkling Water 1.2 for Spark 1.2
- Use Sparkling Water 1.3 for Spark 1.3+
- Use Sparkling Water 1.4 for Spark 1.4
- Use Sparkling Water 1.5 for Spark 1.5
Getting Started with Sparkling Water
Download Sparkling Water: Go here to download Sparkling Water.
Sparkling Water Development Documentation: Read this document first to get started with Sparkling Water.
Launch on Hadoop and Import from HDFS: Go here to learn how to start Sparkling Water on Hadoop.
Sparkling Water Tutorials: Go here for demos and examples.
Sparkling Water K-means Tutorial: Go here to view a demo that uses Scala to create a K-means model.
Sparkling Water GBM Tutorial: Go here to view a demo that uses Scala to create a GBM model.
Sparkling Water on YARN: Follow these instructions to run Sparkling Water on a YARN cluster.
Building Applications on top of H2O: This short tutorial describes project building and demonstrates the capabilities of Sparkling Water using Spark Shell to build a Deep Learning model.
Sparkling Water FAQ: This FAQ provides answers to many common questions about Sparkling Water.
Connecting RStudio to Sparkling Water: This illustrated tutorial describes how to use RStudio to connect to Sparkling Water.
Sparkling Water Blog Posts
Sparkling Water Meetup Slide Decks
PySparkling
Note: PySparkling requires Sparkling Water 1.5 or later.
H2O’s PySparkling package is not available through pip (there is another similarly-named package). H2O’s PySparkling package requires EasyInstall.
To install H2O’s PySparkling package, use the egg file included in the distribution.
- Download Spark 1.5.1.
- Set the
SPARK_HOMEandMASTERvariables as described on the Downloads page. - Download Sparkling Water 1.5
- In the unpacked Sparkling Water directory, run the following command:
easy_install --upgrade sparkling-water-1.5.6/py/dist/pySparkling-1.5.6-py2.7.egg
Python Users
Pythonistas will be glad to know that H2O now provides support for this popular programming language. Python users can also use H2O with IPython notebooks. For more information, refer to the following links.
Click here to view instructions on how to use H2O with Python.
Python readme: This document describes how to setup and install the prerequisites for using Python with H2O.
Python docs: This document represents the definitive guide to using Python with H2O.
Python Parity: This document is is a list of Python capabilities that were previously available only through the H2O R interface but are now available in H2O using the Python interface.
Grid Search in Python: This notebook demonstrates the use of grid search in Python.
R Users
Don’t worry, R users - we still provide R support in the latest version of H2O, just as before. The R components of H2O have been cleaned up, simplified, and standardized, so the command format is easier and more intuitive. Due to these improvements, be aware that any scripts created with previous versions of H2O will need some revision to be compatible with the latest version.
We have provided the following helpful resources to assist R users in upgrading to the latest version, including a document that outlines the differences between versions and a tool that reviews scripts for deprecated or renamed parameters.
Currently, the only version of R that is known to be incompatible with H2O is R version 3.1.0 (codename “Spring Dance”). If you are using that version, we recommend upgrading the R version before using H2O.
To check which version of H2O is installed in R, use versions::installed.versions("h2o").
Click here to view instructions for using H2O with R.
R User Documentation: This document contains all commands in the H2O package for R, including examples and arguments. It represents the definitive guide to using H2O in R.
Porting R Scripts: This document is designed to assist users who have created R scripts using previous versions of H2O. Due to the many improvements in R, scripts created using previous versions of H2O will not work. This document provides a side-by-side comparison of the changes in R for each algorithm, as well as overall structural enhancements R users should be aware of, and provides a link to a tool that assists users in upgrading their scripts.
Connecting RStudio to Sparkling Water: This illustrated tutorial describes how to use RStudio to connect to Sparkling Water.
Ensembles
Ensemble machine learning methods use multiple learning algorithms to obtain better predictive performance.
H2O Ensemble GitHub repository: Location for the H2O Ensemble R package.
Ensemble Documentation: This documentation provides more details on the concepts behind ensembles and how to use them.
API Users
API users will be happy to know that the APIs have been more thoroughly documented in the latest release of H2O and additional capabilities (such as exporting weights and biases for Deep Learning models) have been added.
REST APIs are generated immediately out of the code, allowing users to implement machine learning in many ways. For example, REST APIs could be used to call a model created by sensor data and to set up auto-alerts if the sensor data falls below a specified threshold.
H2O 3 REST API Overview: This document describes how the REST API commands are used in H2O, versioning, experimental APIs, verbs, status codes, formats, schemas, payloads, metadata, and examples.
REST API Reference: This document represents the definitive guide to the H2O REST API.
REST API Schema Reference: This document represents the definitive guide to the H2O REST API schemas.
H2O 3 REST API Overview: This document provides an overview of how APIs are used in H2O, including versioning, URLs, HTTP verbs, status codes, formats, schemas, and examples.
Java Users
For Java developers, the following resources will help you create your own custom app that uses H2O.
H2O Core Java Developer Documentation: The definitive Java API guide for the core components of H2O.
H2O Algos Java Developer Documentation: The definitive Java API guide for the algorithms used by H2O.
h2o-genmodel (POJO) Javadoc: Provides a step-by-step guide to creating and implementing POJOs in a Java application.
SDK Information
The Java API is generated and accessible from the download page.
Developers
If you’re looking to use H2O to help you develop your own apps, the following links will provide helpful references.
For the latest version of IDEA IntelliJ, run ./gradlew idea, then click File > Open within IDEA. Select the .ipr file in the repository and click the Choose button.
For older versions of IDEA IntelliJ, run ./gradlew idea, then Import Project within IDEA and point it to the h2o-3 directory.
Note: This process will take longer, so we recommend using the first method if possible.
For JUnit tests to pass, you may need multiple H2O nodes. Create a “Run/Debug” configuration with the following parameters:
Type: Application
Main class: H2OApp
Use class path of module: h2o-app
After starting multiple “worker” node processes in addition to the JUnit test process, they will cloud up and run the multi-node JUnit tests.
Recommended Systems: This one-page PDF provides a basic overview of the operating systems, languages and APIs, Hadoop resource manager versions, cloud computing environments, browsers, and other resources recommended to run H2O.
Developer Documentation: Detailed instructions on how to build and launch H2O, including how to clone the repository, how to pull from the repository, and how to install required dependencies.
Click here to view instructions on how to use H2O with Maven.
Maven install: This page provides information on how to build a version of H2O that generates the correct IDE files.
apps.h2o.ai: Apps.h2o.ai is designed to support application developers via events, networking opportunities, and a new, dedicated website comprising developer kits and technical specs, news, and product spotlights.
H2O Project Templates: This page provides template info for projects created in Java, Scala, or Sparkling Water.
H2O Scala API Developer Documentation: The definitive Scala API guide for H2O.
Hacking Algos: This blog post by Cliff walks you through building a new algorithm, using K-Means, Quantiles, and Grep as examples.
KV Store Guide: Learn more about performance characteristics when implementing new algorithms.
- Contributing code: If you’re interested in contributing code to H2O, we appreciate your assistance! This document describes how to access our list of Jiras that contributors can work on and how to contact us. Note: To access this link, you must have an Atlassian account.
Downloading H2O
To download H2O, go to our downloads page. Select a build type (bleeding edge or latest alpha), then select an installation method (standalone, R, Python, Hadoop, or Maven) by clicking the tabs at the top of the page. Follow the instructions in the tab to install H2O.
Starting H2O …
There are a variety of ways to start H2O, depending on which client you would like to use.
… From R
To use H2O in R, follow the instructions on the download page.
… From Python
To use H2O in Python, follow the instructions on the download page.
… On Spark
To use H2O on Spark, follow the instructions on the Sparkling Water download page.
… From the Cmd Line
You can use Terminal (OS X) or the Command Prompt (Windows) to launch H2O 3.0. When you launch from the command line, you can include additional instructions to H2O 3.0, such as how many nodes to launch, how much memory to allocate for each node, assign names to the nodes in the cloud, and more.
Note: H2O requires some space in the
/tmpdirectory to launch. If you cannot launch H2O, try freeing up some space in the/tmpdirectory, then try launching H2O again.
For more detailed instructions on how to build and launch H2O, including how to clone the repository, how to pull from the repository, and how to install required dependencies, refer to the developer documentation.
There are two different argument types:
- JVM arguments
- H2O arguments
The arguments use the following format: java <JVM Options> -jar h2o.jar <H2O Options>.
JVM Options
-version: Display Java version info.-Xmx<Heap Size>: To set the total heap size for an H2O node, configure the memory allocation option-Xmx. By default, this option is set to 1 Gb (-Xmx1g). When launching nodes, we recommend allocating a total of four times the memory of your data.
Note: Do not try to launch H2O with more memory than you have available.
H2O Options
-hor-help: Display this information in the command line output.-name <H2OCloudName>: Assign a name to the H2O instance in the cloud (where<H2OCloudName>is the name of the cloud. Nodes with the same cloud name will form an H2O cloud (also known as an H2O cluster).-flatfile <FileName>: Specify a flatfile of IP address for faster cloud formation (where<FileName>is the name of the flatfile.-ip <IPnodeAddress>: Specify an IP address other than the defaultlocalhostfor the node to use (where<IPnodeAddress>is the IP address).-port <#>: Specify a port number other than the default54321for the node to use (where<#>is the port number).-network ###.##.##.#/##: Specify an IP addresses (where###.##.##.#/##represents the IP address and subnet mask). The IP address discovery code binds to the first interface that matches one of the networks in the comma-separated list; to specify an IP address, use-network. To specify a range, use a comma to separate the IP addresses:-network 123.45.67.0/22,123.45.68.0/24. For example,10.1.2.0/24supports 256 possibilities.-ice_root <fileSystemPath>: Specify a directory for H2O to spill temporary data to disk (where<fileSystemPath>is the file path).-flow_dir <server-side or HDFS directory>: Specify a directory for saved flows. The default is/Users/h2o-<H2OUserName>/h2oflows(where<H2OUserName>is your user name).-nthreads <#ofThreads>: Specify the maximum number of threads in the low-priority batch work queue (where<#ofThreads>is the number of threads). The default is 99.-client: Launch H2O node in client mode. This is used mostly for running Sparkling Water.
Cloud Formation Behavior
New H2O nodes join to form a cloud during launch. After a job has started on the cloud, it prevents new members from joining.
To start an H2O node with 4GB of memory and a default cloud name:
java -Xmx4g -jar h2o.jarTo start an H2O node with 6GB of memory and a specific cloud name:
java -Xmx6g -jar h2o.jar -name MyCloudTo start an H2O cloud with three 2GB nodes using the default cloud names:
java -Xmx2g -jar h2o.jar & java -Xmx2g -jar h2o.jar & java -Xmx2g -jar h2o.jar &
Wait for the INFO: Registered: # schemas in: #mS output before entering the above command again to add another node (the number for # will vary).
Flatfile Configuration for Multi-Node Clusters
Running H2O on a multi-node cluster allows you to use more memory for large-scale tasks (for example, creating models from huge datasets) than would be possible on a single node.
If you are configuring many nodes, using the -flatfile option is fast and easy. The -flatfile option is used to define a list of potential cloud peers. However, it is not an alternative to -ip and -port, which should be used to bind the IP and port address of the node you are using to launch H2O.
To configure H2O on a multi-node cluster:
- Locate a set of hosts that will be used to create your cluster. A host can be a server, an EC2 instance, or your laptop.
- Download the appropriate version of H2O for your environment.
- Verify the same h2o.jar file is available on each host in the multi-node cluster.
Create a flatfile.txt that contains an IP address and port number for each H2O instance. Use one entry per line. For example:
192.168.1.163:54321 192.168.1.164:54321- Copy the flatfile.txt to each node in the cluster.
Use the
-Xmxoption to specify the amount of memory for each node. The cluster’s memory capacity is the sum of all H2O nodes in the cluster.For example, if you create a cluster with four 20g nodes (by specifying
-Xmx20gfour times), H2O will have a total of 80 gigs of memory available.For best performance, we recommend sizing your cluster to be about four times the size of your data. To avoid swapping, the
-Xmxallocation must not exceed the physical memory on any node. Allocating the same amount of memory for all nodes is strongly recommended, as H2O works best with symmetric nodes.Note the optional
-ipand-portoptions specify the IP address and ports to use. The-ipoption is especially helpful for hosts with multiple network interfaces.java -Xmx20g -jar h2o.jar -flatfile flatfile.txt -port 54321The output will resemble the following:
04-20 16:14:00.253 192.168.1.70:54321 2754 main INFO: 1. Open a terminal and run 'ssh -L 55555:localhost:54321 H2O-3User@###.###.#.##' 04-20 16:14:00.253 192.168.1.70:54321 2754 main INFO: 2. Point your browser to http://localhost:55555 04-20 16:14:00.437 192.168.1.70:54321 2754 main INFO: Log dir: '/tmp/h2o-H2O-3User/h2ologs' 04-20 16:14:00.437 192.168.1.70:54321 2754 main INFO: Cur dir: '/Users/H2O-3User/h2o-3' 04-20 16:14:00.459 192.168.1.70:54321 2754 main INFO: HDFS subsystem successfully initialized 04-20 16:14:00.460 192.168.1.70:54321 2754 main INFO: S3 subsystem successfully initialized 04-20 16:14:00.460 192.168.1.70:54321 2754 main INFO: Flow dir: '/Users/H2O-3User/h2oflows' 04-20 16:14:00.475 192.168.1.70:54321 2754 main INFO: Cloud of size 1 formed [/192.168.1.70:54321]As you add more nodes to your cluster, the output is updated:
INFO WATER: Cloud of size 2 formed [/...]...Access the H2O 3.0 web UI (Flow) with your browser. Point your browser to the HTTP address specified in the output
Listening for HTTP and REST traffic on ....
On EC2 and S3
On EC2
Tested on Redhat AMI, Amazon Linux AMI, and Ubuntu AMI
To use the Amazon Web Services (AWS) S3 storage solution, you will need to pass your S3 access credentials to H2O. This will allow you to access your data on S3 when importing data frames with path prefixes s3n://....
For security reasons, we recommend writing a script to read the access credentials that are stored in a separate file. This will not only keep your credentials from propagating to other locations, but it will also make it easier to change the credential information later.
Standalone Instance
When running H2O in standalone mode using the simple Java launch command, we can pass in the S3 credentials in two ways.
You can pass in credentials in standalone mode by creating a
core-site.xmlfile and pass it in with the flag-hdfs_config. For an examplecore-site.xmlfile, refer to Core-site.xml.Edit the properties in the core-site.xml file to include your Access Key ID and Access Key as shown in the following example:
<property> <name>fs.s3n.awsAccessKeyId</name> <value>[AWS SECRET KEY]</value> </property> <property> <name>fs.s3n.awsSecretAccessKey</name> <value>[AWS SECRET ACCESS KEY]</value> </property>Launch with the configuration file
core-site.xmlby entering the following in the command line:java -jar h2o.jar -hdfs_config core-site.xml
Import the data using importFile with the S3 url path:
s3n://bucket/path/to/file.csv
You can pass the AWS Access Key and Secret Access Key in an S3N Url in Flow, R, or Python (where
AWS_ACCESS_KEYrepresents your user name andAWS_SECRET_KEYrepresents your password).To import the data from the Flow API:
`importFiles [ "s3n://<AWS_ACCESS_KEY>:<AWS_SECRET_KEY>@bucket/path/to/file.csv" ]`To import the data from the R API:
`h2o.importFile(path = "s3n://<AWS_ACCESS_KEY>:<AWS_SECRET_KEY>@bucket/path/to/file.csv")`To import the data from the Python API:
`h2o.import_file(path = "s3n://<AWS_ACCESS_KEY>:<AWS_SECRET_KEY>@bucket/path/to/file.csv")`
Multi-Node Instance
Python and the
botoPython library are required to launch a multi-node instance of H2O on EC2. Confirm these dependencies are installed before proceeding.
For more information, refer to the H2O EC2 repo.
Build a cluster of EC2 instances by running the following commands on the host that can access the nodes using a public DNS name.
Edit
h2o-cluster-launch-instances.pyto include your SSH key name and security group name, as well as any other environment-specific variables../h2o-cluster-launch-instances.py ./h2o-cluster-distribute-h2o.sh—OR—
./h2o-cluster-launch-instances.py ./h2o-cluster-download-h2o.shNote: The second method may be faster than the first, since download pulls from S3.
Distribute the credentials using
./h2o-cluster-distribute-aws-credentials.sh.Note: If you are running H2O using an IAM role, it is not necessary to distribute the AWS credentials to all the nodes in the cluster. The latest version of H2O can access the temporary access key.
Caution: Distributing the AWS credentials copies the Amazon
AWS_ACCESS_KEY_IDandAWS_SECRET_ACCESS_KEYto the instances to enable S3 and S3N access. Use caution when adding your security keys to the cloud.Start H2O by launching one H2O node per EC2 instance:
./h2o-cluster-start-h2o.shWait 60 seconds after entering the command before entering it on the next node.
In your internet browser, substitute any of the public DNS node addresses for
IP_ADDRESSin the following example:http://IP_ADDRESS:54321- To start H2O:
./h2o-cluster-start-h2o.sh - To stop H2O:
./h2o-cluster-stop-h2o.sh - To shut down the cluster, use your Amazon AWS console to shut down the cluster manually.
Note: To successfully import data, the data must reside in the same location on all nodes.
- To start H2O:
Core-site.xml Example
The following is an example core-site.xml file:
<?xml version="1.0"?>
<?xml-stylesheet type="text/xsl" href="configuration.xsl"?>
<!-- Put site-specific property overrides in this file. -->
<configuration>
<!--
<property>
<name>fs.default.name</name>
<value>s3n://<your s3 bucket></value>
</property>
-->
<property>
<name>fs.s3n.awsAccessKeyId</name>
<value>insert access key here</value>
</property>
<property>
<name>fs.s3n.awsSecretAccessKey</name>
<value>insert secret key here</value>
</property>
</configuration>
Launching H2O
- Selecting the Operating System and Virtualization Type
- Configuring the Instance
- Downloading Java and H2O
Note: Before launching H2O on an EC2 cluster, verify that ports 54321 and 54322 are both accessible by TCP and UDP.
Selecting the Operating System and Virtualization Type
Select your operating system and the virtualization type of the prebuilt AMI on Amazon. If you are using Windows, you will need to use a hardware-assisted virtual machine (HVM). If you are using Linux, you can choose between para-virtualization (PV) and HVM. These selections determine the type of instances you can launch.
For more information about virtualization types, refer to Amazon.
Configuring the Instance
Select the IAM role and policy to use to launch the instance. H2O detects the temporary access keys associated with the instance, so you don’t need to copy your AWS credentials to the instances.
When launching the instance, select an accessible key pair.
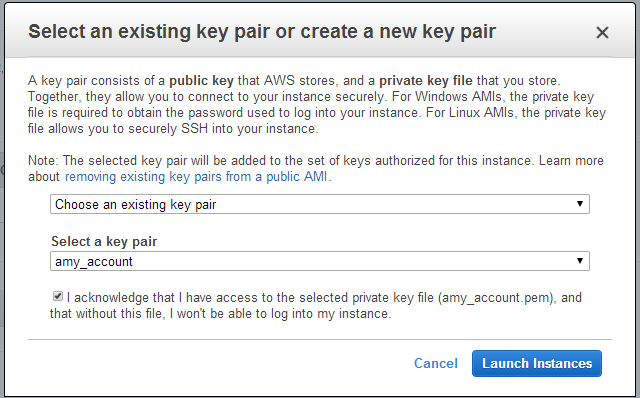
(Windows Users) Tunneling into the Instance
For Windows users that do not have the ability to use ssh from the terminal, either download Cygwin or a Git Bash that has the capability to run ssh:
ssh -i amy_account.pem ec2-user@54.165.25.98
Otherwise, download PuTTY and follow these instructions:
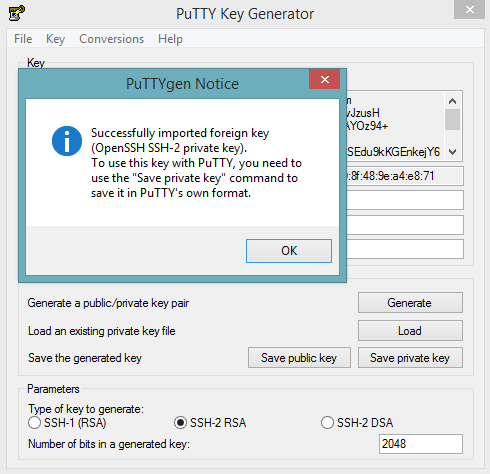
- Launch the PuTTY Key Generator.
- Load your downloaded AWS pem key file. Note: To see the file, change the browser file type to “All”.
Save the private key as a .ppk file.
Launch the PuTTY client.
In the Session section, enter the host name or IP address. For Ubuntu users, the default host name is
ubuntu@<ip-address>. For Linux users, the default host name isec2-user@<ip-address>.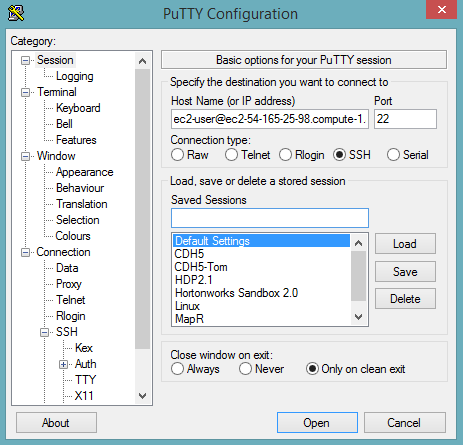
Select SSH, then Auth in the sidebar, and click the Browse button to select the private key file for authentication.
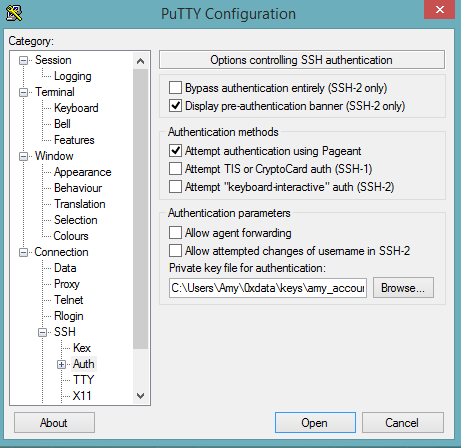
Start a new session and click the Yes button to confirm caching of the server’s rsa2 key fingerprint and continue connecting.
Downloading Java and H2O
- Download Java (JDK 1.7 or later) if it is not already available on the instance.
To download H2O, run the
wgetcommand with the link to the zip file available on our website by copying the link associated with the Download button for the selected H2O build.wget http://h2o-release.s3.amazonaws.com/h2o/rel-turchin/3/index.html unzip h2o-3.8.2.3.zip cd h2o-3.8.2.3 java -Xmx4g -jar h2o.jarFrom your browser, navigate to
<Private_IP_Address>:54321or<Public_DNS>:54321to use H2O’s web interface.
… On Hadoop
Currently supported versions:
- CDH 5.2
- CDH 5.3
- CDH 5.4.2
- HDP 2.1
- HDP 2.2
- HDP 2.3
- MapR 3.1.1
- MapR 4.0.1
- MapR 5.0
Important Points to Remember:
- The command used to launch H2O differs from previous versions (refer to the Tutorial section)
- Launching H2O on Hadoop requires at least 6 GB of memory
- Each H2O node runs as a mapper
- Run only one mapper per host
- There are no combiners or reducers
- Each H2O cluster must have a unique job name
-mapperXmx,-nodes, and-outputare required- Root permissions are not required - just unzip the H2O .zip file on any single node
Prerequisite: Open Communication Paths
H2O communicates using two communication paths. Verify these are open and available for use by H2O.
Path 1: mapper to driver
Optionally specify this port using the -driverport option in the hadoop jar command (see “Hadoop Launch Parameters” below). This port is opened on the driver host (the host where you entered the hadoop jar command). By default, this port is chosen randomly by the operating system.
Path 2: mapper to mapper
Optionally specify this port using the -baseport option in the hadoop jar command (refer to Hadoop Launch Parameters below. This port and the next subsequent port are opened on the mapper hosts (the Hadoop worker nodes) where the H2O mapper nodes are placed by the Resource Manager. By default, ports 54321 (TCP) and 54322 (TCP & UDP) are used.
The mapper port is adaptive: if 54321 and 54322 are not available, H2O will try 54323 and 54324 and so on. The mapper port is designed to be adaptive because sometimes if the YARN cluster is low on resources, YARN will place two H2O mappers for the same H2O cluster request on the same physical host. For this reason, we recommend opening a range of more than two ports (20 ports should be sufficient).
Tutorial
The following tutorial will walk the user through the download or build of H2O and the parameters involved in launching H2O from the command line.
Download the latest H2O release for your version of Hadoop:
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-cdh5.2.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-cdh5.3.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-hdp2.1.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-hdp2.2.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-hdp2.3.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-mapr3.1.1.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-mapr4.0.1.zip wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-mapr5.0.zipNote: Enter only one of the above commands.
Prepare the job input on the Hadoop Node by unzipping the build file and changing to the directory with the Hadoop and H2O’s driver jar files.
unzip h2o-3.8.2.3-*.zip cd h2o-3.8.2.3-*To launch H2O nodes and form a cluster on the Hadoop cluster, run:
hadoop jar h2odriver.jar -nodes 1 -mapperXmx 6g -output hdfsOutputDirNameThe above command launches a 6g node of H2O. We recommend you launch the cluster with at least four times the memory of your data file size.
mapperXmx is the mapper size or the amount of memory allocated to each node. Specify at least 6 GB.
nodes is the number of nodes requested to form the cluster.
output is the name of the directory created each time a H2O cloud is created so it is necessary for the name to be unique each time it is launched.
To monitor your job, direct your web browser to your standard job tracker Web UI. To access H2O’s Web UI, direct your web browser to one of the launched instances. If you are unsure where your JVM is launched, review the output from your command after the nodes has clouded up and formed a cluster. Any of the nodes’ IP addresses will work as there is no master node.
Determining driver host interface for mapper->driver callback... [Possible callback IP address: 172.16.2.181] [Possible callback IP address: 127.0.0.1] ... Waiting for H2O cluster to come up... H2O node 172.16.2.184:54321 requested flatfile Sending flatfiles to nodes... [Sending flatfile to node 172.16.2.184:54321] H2O node 172.16.2.184:54321 reports H2O cluster size 1 H2O cluster (1 nodes) is up Blocking until the H2O cluster shuts down...
Hadoop Launch Parameters
-h | -help: Display help-jobname <JobName>: Specify a job name for the Jobtracker to use; the default isH2O_nnnnn(where n is chosen randomly)-driverif <IP address of mapper -> driver callback interface>: Specify the IP address for callback messages from the mapper to the driver.-driverport <port of mapper -> callback interface>: Specify the port number for callback messages from the mapper to the driver.-network <IPv4Network1>[,<IPv4Network2>]: Specify the IPv4 network(s) to bind to the H2O nodes; multiple networks can be specified to force H2O to use the specified host in the Hadoop cluster.10.1.2.0/24allows 256 possibilities.-timeout <seconds>: Specify the timeout duration (in seconds) to wait for the cluster to form before failing. Note: The default value is 120 seconds; if your cluster is very busy, this may not provide enough time for the nodes to launch. If H2O does not launch, try increasing this value (for example,-timeout 600).-disown: Exit the driver after the cluster forms.-notify <notification file name>: Specify a file to write when the cluster is up. The file contains the IP and port of the embedded web server for one of the nodes in the cluster. All mappers must start before the H2O cloud is considered “up”.-mapperXmx <per mapper Java Xmx heap size>: Specify the amount of memory to allocate to H2O (at least 6g).-extramempercent <0-20>: Specify the extra memory for internal JVM use outside of the Java heap. This is a percentage ofmapperXmx.-n | -nodes <number of H2O nodes>: Specify the number of nodes.-nthreads <maximum number of CPUs>: Specify the number of CPUs to use. Enter-1to use all CPUs on the host, or enter a positive integer.-baseport <initialization port for H2O nodes>: Specify the initialization port for the H2O nodes. The default is54321.-ea: Enable assertions to verify boolean expressions for error detection.-verbose:gc: Include heap and garbage collection information in the logs.-XX:+PrintGCDetails: Include a short message after each garbage collection.-license <license file name>: Specify the directory of local filesytem location and the license file name.-o | -output <HDFS output directory>: Specify the HDFS directory for the output.-flow_dir <Saved Flows directory>: Specify the directory for saved flows. By default, H2O will try to find the HDFS home directory to use as the directory for flows. If the HDFS home directory is not found, flows cannot be saved unless a directory is specified using-flow_dir.
Accessing S3 Data from Hadoop
H2O launched on Hadoop can access S3 Data in addition to to HDFS. To enable access, follow the instructions below.
Edit Hadoop’s core-site.xml, then set the HADOOP_CONF_DIR environment property to the directory containing the core-site.xml file. For an example core-site.xml file, refer to Core-site.xml. Typically, the configuration directory for most Hadoop distributions is /etc/hadoop/conf.
You can also pass the S3 credentials when launching H2O with the Hadoop jar command. Use the -D flag to pass the credentials:
hadoop jar h2odriver.jar -Dfs.s3.awsAccessKeyId="${AWS_ACCESS_KEY}" -Dfs.s3n.awsSecretAccessKey="${AWS_SECRET_KEY}" -n 3 -mapperXmx 10g -output outputDirectory
where AWS_ACCESS_KEY represents your user name and AWS_SECRET_KEY represents your password.
Then import the data with the S3 URL path:
To import the data from the Flow API:
importFiles [ "s3n:/path/to/bucket/file/file.tab.gz" ]To import the data from the R API:
h2o.importFile(path = "s3n://bucket/path/to/file.csv")To import the data from the Python API:
h2o.import_frame(path = "s3n://bucket/path/to/file.csv")
… Using Docker
This walkthrough describes:
- Installing Docker on Mac or Linux OS
- Creating and modifying the Dockerfile
- Building a Docker image from the Dockerfile
- Running the Docker build
- Launching H2O
- Accessing H2O from the web browser or R
Walkthrough
Prerequisites
Linux kernel version 3.8+
or
Mac OS X 10.6+
- VirtualBox
- Latest version of Docker is installed and configured
- Docker daemon is running - enter all commands below in the Docker daemon window
- Using
Userdirectory (notroot)
Notes
- Older Linux kernel versions are known to cause kernel panics that break Docker; there are ways around it, but these should be attempted at your own risk. To check the version of your kernel, run
uname -rat the command prompt. The following walkthrough has been tested on a Mac OS X 10.10.1. - The Dockerfile always pulls the latest H2O release.
- The Docker image only needs to be built once.
Step 1 - Install and Launch Docker
Depending on your OS, select the appropriate installation method:
Step 2 - Create or Download Dockerfile
Note: If the following commands do not work, prepend with
sudo.
Create a folder on the Host OS to host your Dockerfile by running:
mkdir -p /data/h2o-rel-turchin
Next, either download or create a Dockerfile, which is a build recipe that builds the container.
Download and use our Dockerfile template by running:
cd /data/h2o-rel-turchin
wget https://raw.githubusercontent.com/h2oai/h2o-3/master/Dockerfile
The Dockerfile:
- obtains and updates the base image (Ubuntu 14.04)
- installs Java 7
- obtains and downloads the H2O build from H2O’s S3 repository
- exposes ports 54321 and 54322 in preparation for launching H2O on those ports
Step 3 - Build Docker image from Dockerfile
From the /data/h2o-rel-turchin directory, run:
docker build -t "h2oai/rel-turchin:v5" .
Note:
v5represents the current version number.
Because it assembles all the necessary parts for the image, this process can take a few minutes.
Step 4 - Run Docker Build
On a Mac, use the argument -p 54321:54321 to expressly map the port 54321. This is not necessary on Linux.
docker run -ti -p 54321:54321 h2o.ai/rel-turchin:v5 /bin/bash
Note:
v5represents the version number.
Step 5 - Launch H2O
Navigate to the /opt directory and launch H2O. Change the value of -Xmx to the amount of memory you want to allocate to the H2O instance. By default, H2O launches on port 54321.
cd /opt
java -Xmx1g -jar h2o.jar
Step 6 - Access H2O from the web browser or R
- On Linux: After H2O launches, copy and paste the IP address and port of the H2O instance into the address bar of your browser. In the following example, the IP is
172.17.0.5:54321.
03:58:25.963 main INFO WATER: Cloud of size 1 formed [/172.17.0.5:54321 (00:00:00.000)]
- On OSX: Locate the IP address of the Docker’s network (
192.168.59.103in the following examples) that bridges to your Host OS by opening a new Terminal window (not a bash for your container) and runningboot2docker ip.
$ boot2docker ip
192.168.59.103
You can also view the IP address (192.168.99.100 in the example below) by scrolling to the top of the Docker daemon window:
## .
## ## ## ==
## ## ## ## ## ===
/"""""""""""""""""\___/ ===
~~~ {~~ ~~~~ ~~~ ~~~~ ~~~ ~ / ===- ~~~
\______ o __/
\ \ __/
\____\_______/
docker is configured to use the default machine with IP 192.168.99.100
For help getting started, check out the docs at https://docs.docker.com
After obtaining the IP address, point your browser to the specified ip address and port. In R, you can access the instance by installing the latest version of the H2O R package and running:
library(h2o)
dockerH2O <- h2o.init(ip = "192.168.59.103", port = 54321)
Flow Web UI …
H2O Flow is an open-source user interface for H2O. It is a web-based interactive environment that allows you to combine code execution, text, mathematics, plots, and rich media in a single document.
With H2O Flow, you can capture, rerun, annotate, present, and share your workflow. H2O Flow allows you to use H2O interactively to import files, build models, and iteratively improve them. Based on your models, you can make predictions and add rich text to create vignettes of your work - all within Flow’s browser-based environment.
Flow’s hybrid user interface seamlessly blends command-line computing with a modern graphical user interface. However, rather than displaying output as plain text, Flow provides a point-and-click user interface for every H2O operation. It allows you to access any H2O object in the form of well-organized tabular data.
H2O Flow sends commands to H2O as a sequence of executable cells. The cells can be modified, rearranged, or saved to a library. Each cell contains an input field that allows you to enter commands, define functions, call other functions, and access other cells or objects on the page. When you execute the cell, the output is a graphical object, which can be inspected to view additional details.
While H2O Flow supports REST API, R scripts, and CoffeeScript, no programming experience is required to run H2O Flow. You can click your way through any H2O operation without ever writing a single line of code. You can even disable the input cells to run H2O Flow using only the GUI. H2O Flow is designed to guide you every step of the way, by providing input prompts, interactive help, and example flows.
Introduction
This guide will walk you through how to use H2O’s web UI, H2O Flow. To view a demo video of H2O Flow, click here.
Getting Help
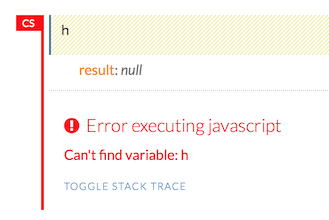
First, let’s go over the basics. Type h to view a list of helpful shortcuts.
The following help window displays:

To close this window, click the X in the upper-right corner, or click the Close button in the lower-right corner. You can also click behind the window to close it. You can also access this list of shortcuts by clicking the Help menu and selecting Keyboard Shortcuts.
For additional help, click Help > Assist Me or click the Assist Me! button in the row of buttons below the menus.

You can also type assist in a blank cell and press Ctrl+Enter. A list of common tasks displays to help you find the correct command.
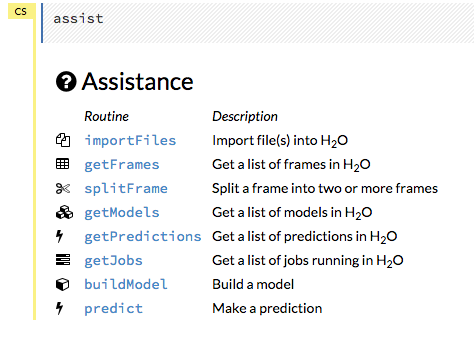
There are multiple resources to help you get started with Flow in the Help sidebar.
Note: To hide the sidebar, click the >> button above it.
To display the sidebar if it is hidden, click the << button.
To access this documentation, select the Flow Web UI… link below the General heading in the Help sidebar.
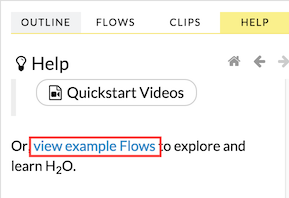
You can also explore the pre-configured flows available in H2O Flow for a demonstration of how to create a flow. To view the example flows:
Click the view example Flows link below the Quickstart Videos button in the Help sidebar
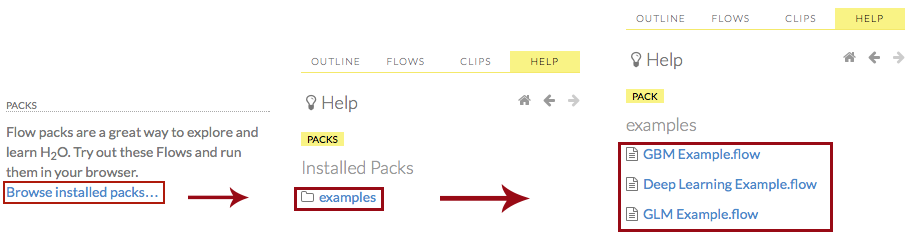
or
Click the Browse installed packs… link in the Packs subsection of the Help sidebar. Click the examples folder and select the example flow from the list.
If you have a flow currently open, a confirmation window appears asking if the current notebook should be replaced. To load the example flow, click the Load Notebook button.
To view the REST API documentation, click the Help tab in the sidebar and then select the type of REST API documentation (Routes or Schemas).

Before getting started with H2O Flow, make sure you understand the different cell modes. Certain actions can only be performed when the cell is in a specific mode.
Understanding Cell Modes
- Using Edit Mode
- Using Command Mode
- Changing Cell Formats
- Running Cells
- Running Flows
- Using Keyboard Shortcuts
- Using Variables in Cells
- Using Flow Buttons
There are two modes for cells: edit and command.
Using Edit Mode
In edit mode, the cell is yellow with a blinking bar to indicate where text can be entered and there is an orange flag to the left of the cell.

Using Command Mode
In command mode, the flag is yellow. The flag also indicates the cell’s format:
MD: Markdown
Note: Markdown formatting is not applied until you run the cell by:
clicking the Run button
 or
orpressing Ctrl+Enter

CS: Code (default)

RAW: Raw format (for code comments)

H[1-6]: Heading level (where 1 is a first-level heading)

NOTE: If there is an error in the cell, the flag is red.
If the cell is executing commands, the flag is teal. The flag returns to yellow when the task is complete.
Changing Cell Formats
To change the cell’s format (for example, from code to Markdown), make sure you are in command (not edit) mode and that the cell you want to change is selected. The easiest way to do this is to click on the flag to the left of the cell. Enter the keyboard shortcut for the format you want to use. The flag’s text changes to display the current format.
| Cell Mode | Keyboard Shortcut |
|---|---|
| Code | y |
| Markdown | m |
| Raw text | r |
| Heading 1 | 1 |
| Heading 2 | 2 |
| Heading 3 | 3 |
| Heading 4 | 4 |
| Heading 5 | 5 |
| Heading 6 | 6 |
Running Cells
The series of buttons at the top of the page below the menus run cells in a flow.
- To run all cells in the flow, click the Flow menu, then click Run All Cells.
- To run the current cell and all subsequent cells, click the Flow menu, then click Run All Cells Below.
To run an individual cell in a flow, confirm the cell is in Edit Mode, then:
press Ctrl+Enter
or
click the Run button

Running Flows
When you run the flow, a progress bar indicates the current status of the flow. You can cancel the currently running flow by clicking the Stop button in the progress bar.
When the flow is complete, a message displays in the upper right.
Note: If there is an error in the flow, H2O Flow stops at the cell that contains the error.
Using Keyboard Shortcuts
Here are some important keyboard shortcuts to remember:
- Click a cell and press Enter to enter edit mode, which allows you to change the contents of a cell.
- To exit edit mode, press Esc.
- To execute the contents of a cell, press the Ctrl and Enter buttons at the same time.
The following commands must be entered in command mode.
- To add a new cell above the current cell, press a.
- To add a new cell below the current cell, press b.
- To delete the current cell, press the d key twice. (dd).
You can view these shortcuts by clicking Help > Keyboard Shortcuts or by clicking the Help tab in the sidebar.
Using Variables in Cells
Variables can be used to store information such as download locations. To use a variable in Flow:
- Define the variable in a code cell (for example,
locA = "https://h2o-public-test-data.s3.amazonaws.com/bigdata/laptop/kdd2009/small-churn/kdd_train.csv").
- Run the cell. H2O validates the variable.

- Use the variable in another code cell (for example,
importFiles [locA]).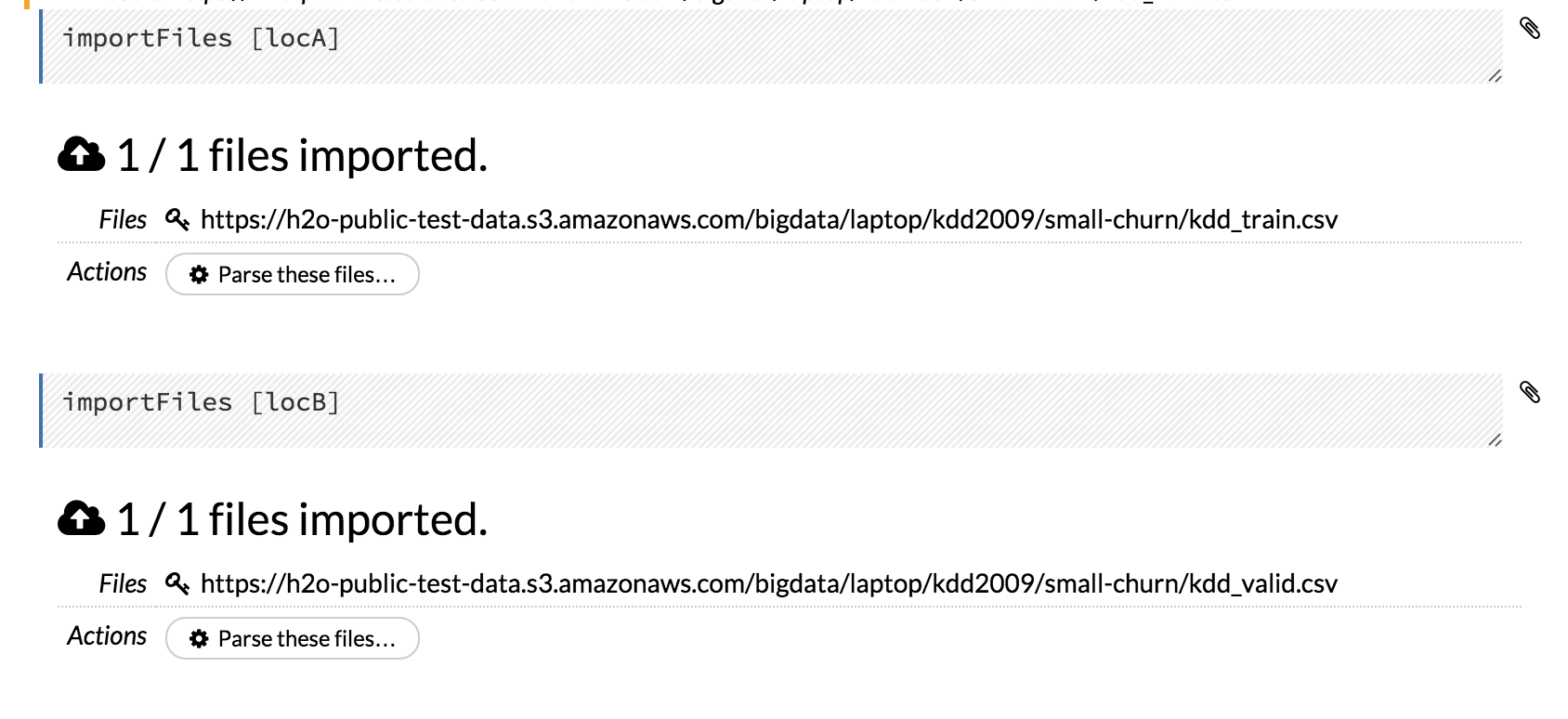 To further simplify your workflow, you can save the cells containing the variables and definitions as clips.
To further simplify your workflow, you can save the cells containing the variables and definitions as clips.
Using Flow Buttons
There are also a series of buttons at the top of the page below the flow name that allow you to save the current flow, add a new cell, move cells up or down, run the current cell, and cut, copy, or paste the current cell. If you hover over the button, a description of the button’s function displays.

You can also use the menus at the top of the screen to edit the order of the cells, toggle specific format types (such as input or output), create models, or score models. You can also access troubleshooting information or obtain help with Flow.

Note: To disable the code input and use H2O Flow strictly as a GUI, click the Cell menu, then Toggle Cell Input.
Now that you are familiar with the cell modes, let’s import some data.
… Importing Data
If you don’t have any data of your own to work with, you can find some example datasets here:
There are multiple ways to import data in H2O flow:
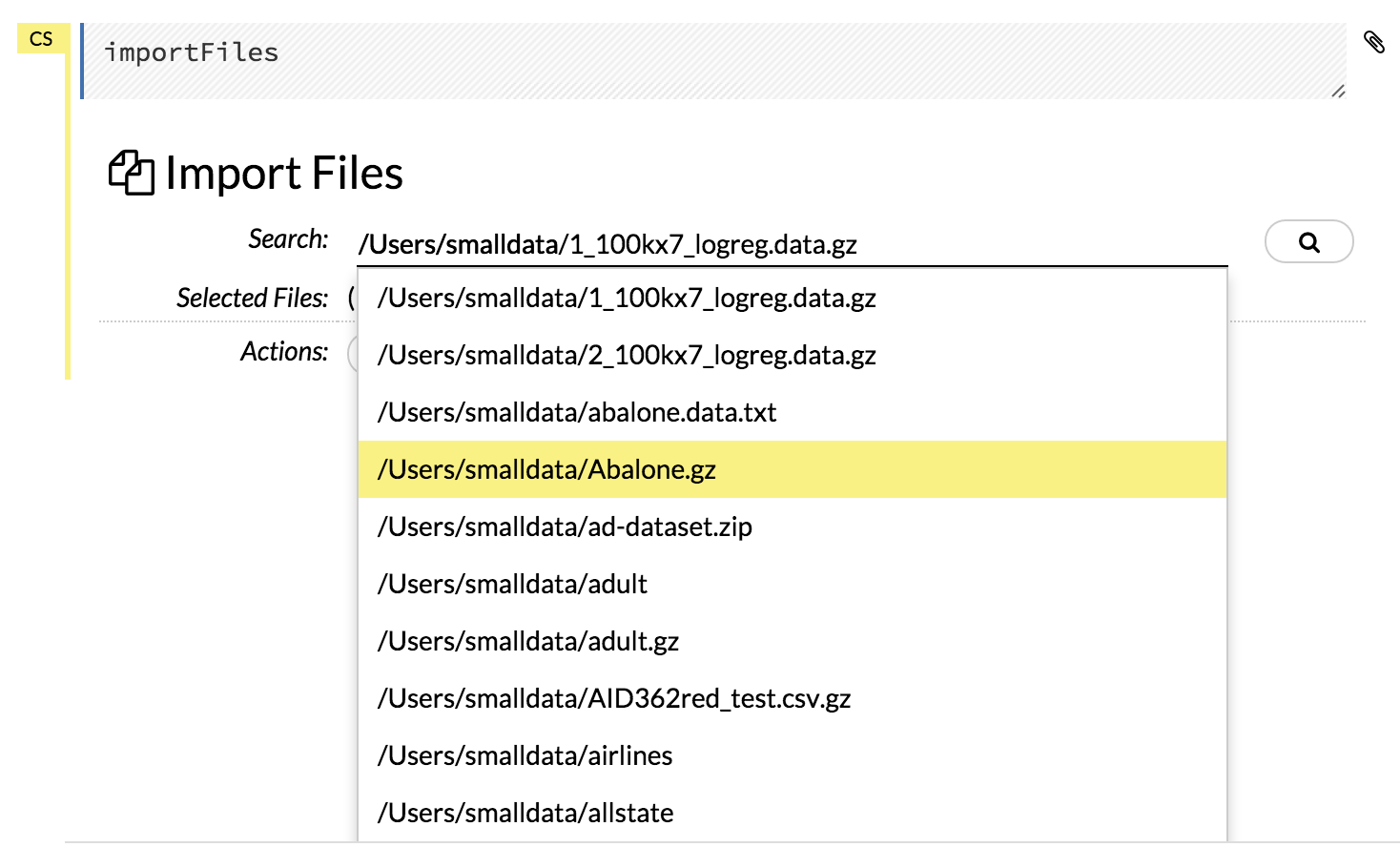
Click the Assist Me! button in the row of buttons below the menus, then click the importFiles link. Enter the file path in the auto-completing Search entry field and press Enter. Select the file from the search results and confirm it by clicking the Add All link.
In a blank cell, select the CS format, then enter
importFiles ["path/filename.format"](wherepath/filename.formatrepresents the complete file path to the file, including the full file name. The file path can be a local file path or a website address.
Note: For S3 file locations, use the format
importFiles [ "s3n:/path/to/bucket/file/file.tab.gz" ]
- For an example of how to import a single file or a directory in R, refer to the following example.
After selecting the file to import, the file path displays in the “Search Results” section. To import a single file, click the plus sign next to the file. To import all files in the search results, click the Add all link. The files selected for import display in the “Selected Files” section.
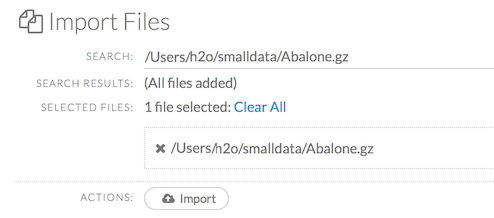
Note: If the file is compressed, it will only be read using a single thread. For best performance, we recommend uncompressing the file before importing, as this will allow use of the faster multithreaded distributed parallel reader during import. Please note that .zip files containing multiple files are not currently supported.
To import the selected file(s), click the Import button.
To remove all files from the “Selected Files” list, click the Clear All link.
To remove a specific file, click the X next to the file path.
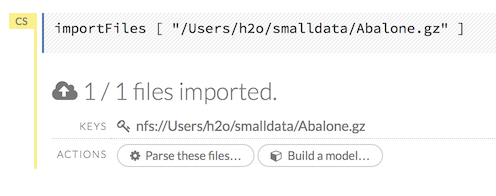
After you click the Import button, the raw code for the current job displays. A summary displays the results of the file import, including the number of imported files and their Network File System (nfs) locations.
Uploading Data
To upload a local file, click the Data menu and select Upload File…. Click the Choose File button, select the file, click the Choose button, then click the Upload button.
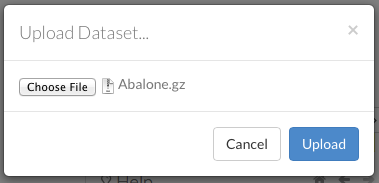
When the file has uploaded successfully, a message displays in the upper right and the Setup Parse cell displays.

Ok, now that your data is available in H2O Flow, let’s move on to the next step: parsing. Click the Parse these files button to continue.
Parsing Data
After you have imported your data, parse the data.
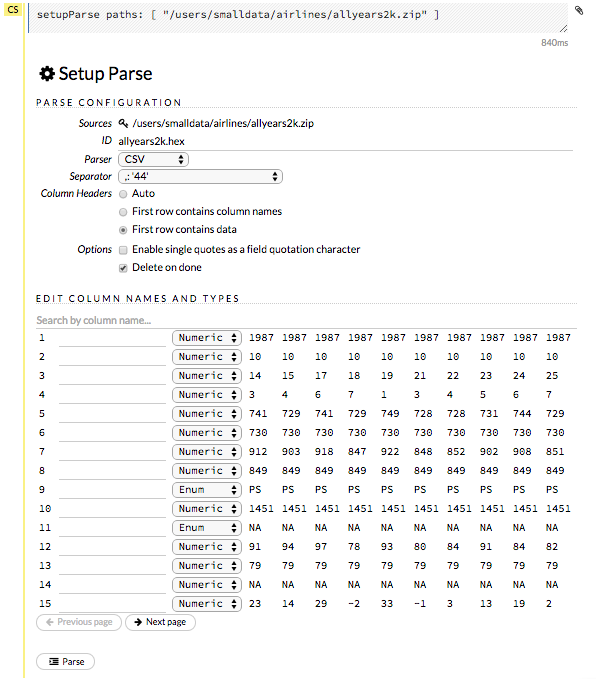
The read-only Sources field displays the file path for the imported data selected for parsing.
The ID contains the auto-generated name for the parsed data (by default, the file name of the imported file with .hex as the file extension). Use the default name or enter a custom name in this field.
Select the parser type (if necessary) from the drop-down Parser list. For most data parsing, H2O automatically recognizes the data type, so the default settings typically do not need to be changed. The following options are available:
- Auto
- ARFF
- XLS
- XLSX
- CSV
SVMLight
Note: For SVMLight data, the column indices must be >= 1 and the columns must be in ascending order.
If a separator or delimiter is used, select it from the Separator list.
Select a column header option, if applicable:
- Auto: Automatically detect header types.
- First row contains column names: Specify heading as column names.
- First row contains data: Specify heading as data. This option is selected by default.
Select any necessary additional options:
- Enable single quotes as a field quotation character: Treat single quote marks (also known as apostrophes) in the data as a character, rather than an enum. This option is not selected by default.
- Delete on done: Check this checkbox to delete the imported data after parsing. This option is selected by default.
A preview of the data displays in the “Edit Column Names and Types” section.
To change or add a column name, edit or enter the text in the column’s entry field. In the screenshot below, the entry field for column 16 is highlighted in red.
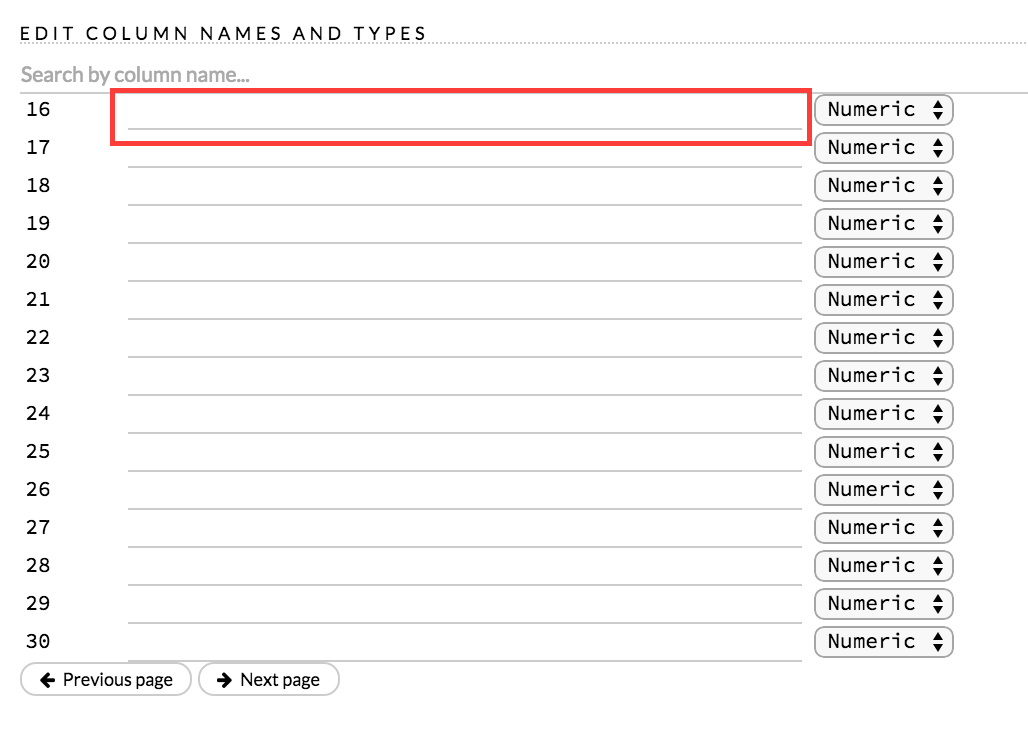
To change the column type, select the drop-down list to the right of the column name entry field and select the data type. The options are:
- Unknown
- Numeric
- Enum
- Time
- UUID
- String
- Invalid
You can search for a column by entering it in the Search by column name… entry field above the first column name entry field. As you type, H2O displays the columns that match the specified search terms.
Note: Only custom column names are searchable. Default column names cannot be searched.
To navigate the data preview, click the <- Previous page or -> Next page buttons.
After making your selections, click the Parse button.
After you click the Parse button, the code for the current job displays.

Since we’ve submitted a couple of jobs (data import & parse) to H2O now, let’s take a moment to learn more about jobs in H2O.
Viewing Jobs
Any command (such as importFiles) you enter in H2O is submitted as a job, which is associated with a key. The key identifies the job within H2O and is used as a reference.
Viewing All Jobs
To view all jobs, click the Admin menu, then click Jobs, or enter getJobs in a cell in CS mode.
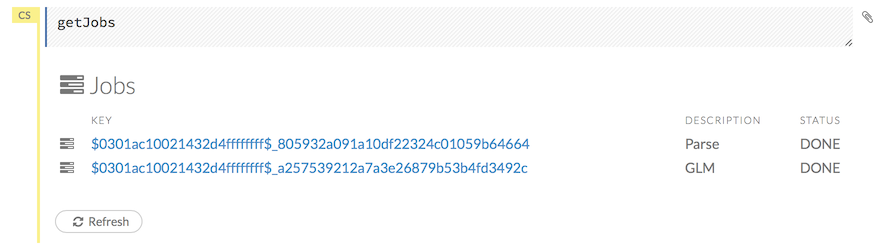
The following information displays:
- Type (for example,
FrameorModel) - Link to the object
- Description of the job type (for example,
ParseorGBM) - Start time
- End time
- Run time
To refresh this information, click the Refresh button. To view the details of the job, click the View button.
Viewing Specific Jobs
To view a specific job, click the link in the “Destination” column.
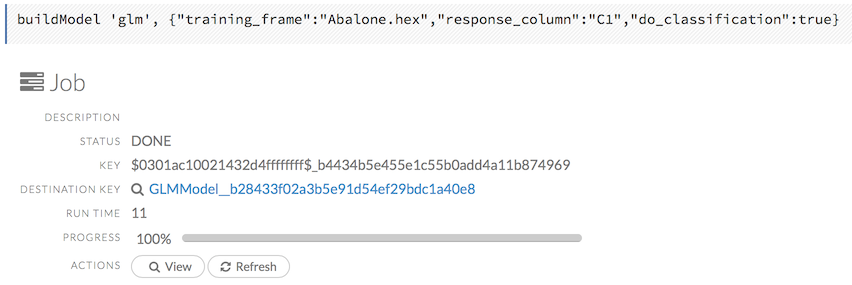
The following information displays:
- Type (for example,
Frame) - Link to object (key)
- Description (for example,
Parse) - Status
- Run time
- Progress
NOTE: For a better understanding of how jobs work, make sure to review the Viewing Frames section as well.
Ok, now that you understand how to find jobs in H2O, let’s submit a new one by building a model.
… Building Models
To build a model:
Click the Assist Me! button in the row of buttons below the menus and select buildModel
or
Click the Assist Me! button, select getFrames, then click the Build Model… button below the parsed .hex data set
or
Click the View button after parsing data, then click the Build Model button
or
Click the drop-down Model menu and select the model type from the list
The Build Model… button can be accessed from any page containing the .hex key for the parsed data (for example, getJobs > getFrame). The following image depicts the K-Means model type. Available options vary depending on model type.
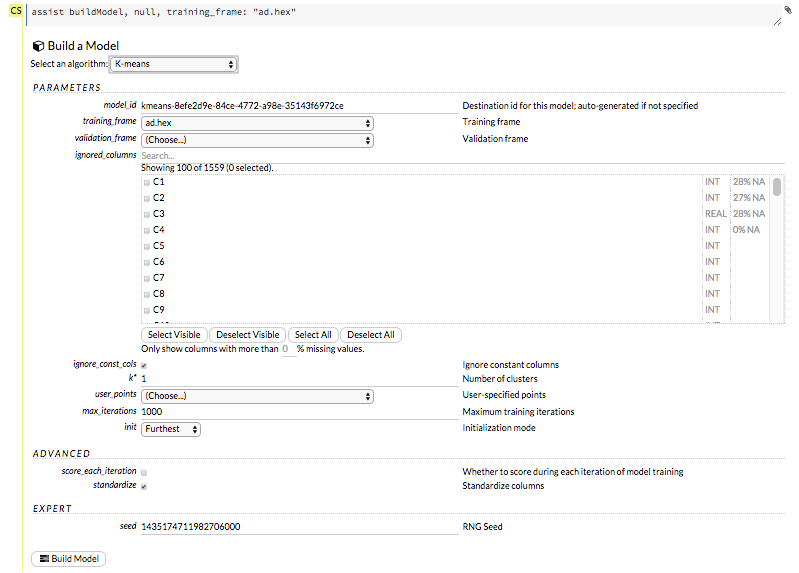
In the Build a Model cell, select an algorithm from the drop-down menu:
- K-means: Create a K-Means model.
- Generalized Linear Model: Create a Generalized Linear model.
- Distributed RF: Create a distributed Random Forest model.
- Naïve Bayes: Create a Naïve Bayes model.
- Principal Component Analysis: Create a Principal Components Analysis model for modeling without regularization or performing dimensionality reduction.
- Gradient Boosting Machine: Create a Gradient Boosted model
- Deep Learning: Create a Deep Learning model.
The available options vary depending on the selected model. If an option is only available for a specific model type, the model type is listed. If no model type is specified, the option is applicable to all model types.
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates an ID containing the model type (for example,
gbm-6f6bdc8b-ccbc-474a-b590-4579eea44596).training_frame: (Required) Select the dataset used to build the model.
validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
nfolds: (GLM, GBM, DL, DRF) Specify the number of folds for cross-validation.
response_column: (Required for GLM, GBM, DL, DRF, Naïve Bayes) Select the column to use as the independent variable.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: (Optional) Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
transform: (PCA) Select the transformation method for the training data: None, Standardize, Normalize, Demean, or Descale.
pca_method: (PCA) Select the algorithm to use for computing the principal components:
- GramSVD: Uses a distributed computation of the Gram matrix, followed by a local SVD using the JAMA package
- Power: Computes the SVD using the power iteration method
- Randomized: Uses randomized subspace iteration method
- GLRM: Fits a generalized low-rank model with L2 loss function and no regularization and solves for the SVD using local matrix algebra
family: (GLM) Select the model type (Gaussian, Binomial, Multinomial, Poisson, Gamma, or Tweedie).
solver: (GLM) Select the solver to use (AUTO, IRLSM, L_BFGS, COORDINATE_DESCENT_NAIVE, or COORDINATE_DESCENT). IRLSM is fast on on problems with a small number of predictors and for lambda-search with L1 penalty, while L_BFGS scales better for datasets with many columns. COORDINATE_DESCENT is IRLSM with the covariance updates version of cyclical coordinate descent in the innermost loop. COORDINATE_DESCENT_NAIVE is IRLSM with the naive updates version of cyclical coordinate descent in the innermost loop. COORDINATE_DESCENT_NAIVE and COORDINATE_DESCENT are currently experimental.
link: (GLM) Select a link function (Identity, Family_Default, Logit, Log, Inverse, or Tweedie).
alpha: (GLM) Specify the regularization distribution between L2 and L2.
lambda: (GLM) Specify the regularization strength.
lambda_search: (GLM) Check this checkbox to enable lambda search, starting with lambda max. The given lambda is then interpreted as lambda min.
non-negative: (GLM) To force coefficients to be non-negative, check this checkbox.
standardize: (K-Means, GLM) To standardize the numeric columns to have mean of zero and unit variance, check this checkbox. Standardization is highly recommended; if you do not use standardization, the results can include components that are dominated by variables that appear to have larger variances relative to other attributes as a matter of scale, rather than true contribution. This option is selected by default.
beta_constraints: (GLM) To use beta constraints, select a dataset from the drop-down menu. The selected frame is used to constraint the coefficient vector to provide upper and lower bounds.
min_rows: (GBM, DRF) Specify the minimum number of observations for a leaf (“nodesize” in R).
nbins: (GBM, DRF) (Numerical [real/int] only) Specify the minimum number of bins for the histogram to build, then split at the best point.
nbins_cats: (GBM, DRF) (Categorical [factors/enums] only) Specify the maximum number of bins for the histogram to build, then split at the best point. Higher values can lead to more overfitting. The levels are ordered alphabetically; if there are more levels than bins, adjacent levels share bins. This value has a more significant impact on model fitness than nbins. Larger values may increase runtime, especially for deep trees and large clusters, so tuning may be required to find the optimal value for your configuration.
learn_rate: (GBM) Specify the learning rate. The range is 0.0 to 1.0.
distribution: (GBM, DL) Select the distribution type from the drop-down list. The options are auto, bernoulli, multinomial, gaussian, poisson, gamma, or tweedie.
sample_rate: (GBM, DRF) Specify the row sampling rate (x-axis). The range is 0.0 to 1.0. Higher values may improve training accuracy. Test accuracy improves when either columns or rows are sampled. For details, refer to “Stochastic Gradient Boosting” (Friedman, 1999).
col_sample_rate: (GBM, DRF) Specify the column sampling rate (y-axis). The range is 0.0 to 1.0. Higher values may improve training accuracy. Test accuracy improves when either columns or rows are sampled. For details, refer to “Stochastic Gradient Boosting” (Friedman, 1999).
mtries: (DRF) Specify the columns to randomly select at each level. If the default value of
-1is used, the number of variables is the square root of the number of columns for classification and p/3 for regression (where p is the number of predictors).binomial_double_trees: (DRF) (Binary classification only) Build twice as many trees (one per class). Enabling this option can lead to higher accuracy, while disabling can result in faster model building. This option is disabled by default.
score_each_iteration: (K-Means, DRF, Naïve Bayes, PCA, GBM, GLM) To score during each iteration of the model training, check this checkbox.
k*: (K-Means, PCA) For K-Means, specify the number of clusters. For PCA, specify the rank of matrix approximation.
user_points: (K-Means) For K-Means, specify the number of initial cluster centers.
max_iterations: (K-Means, PCA, GLM) Specify the number of training iterations.
init: (K-Means) Select the initialization mode. The options are Furthest, PlusPlus, Random, or User.
Note: If PlusPlus is selected, the initial Y matrix is chosen by the final cluster centers from the K-Means PlusPlus algorithm.
tweedie_variance_power: (GLM) (Only applicable if Tweedie is selected for Family) Specify the Tweedie variance power.
tweedie_link_power: (GLM) (Only applicable if Tweedie is selected for Family) Specify the Tweedie link power.
activation: (DL) Select the activation function (Tanh, TanhWithDropout, Rectifier, RectifierWithDropout, Maxout, MaxoutWithDropout). The default option is Rectifier.
hidden: (DL) Specify the hidden layer sizes (e.g., 100,100). For Grid Search, use comma-separated values: (10,10),(20,20,20). The default value is [200,200]. The specified value(s) must be positive.
epochs: (DL) Specify the number of times to iterate (stream) the dataset. The value can be a fraction.
variable_importances: (DL) Check this checkbox to compute variable importance. This option is not selected by default.
laplace: (Naïve Bayes) Specify the Laplace smoothing parameter.
min_sdev: (Naïve Bayes) Specify the minimum standard deviation to use for observations without enough data.
eps_sdev: (Naïve Bayes) Specify the threshold for standard deviation. If this threshold is not met, the min_sdev value is used.
min_prob: (Naïve Bayes) Specify the minimum probability to use for observations without enough data.
eps_prob: (Naïve Bayes) Specify the threshold for standard deviation. If this threshold is not met, the min_sdev value is used.
compute_metrics: (Naïve Bayes, PCA) To compute metrics on training data, check this checkbox. The Naïve Bayes classifier assumes independence between predictor variables conditional on the response, and a Gaussian distribution of numeric predictors with mean and standard deviation computed from the training dataset. When building a Naïve Bayes classifier, every row in the training dataset that contains at least one NA will be skipped completely. If the test dataset has missing values, then those predictors are omitted in the probability calculation during prediction.
Advanced Options
fold_assignment: (GLM, GBM, DL, DRF, K-Means) (Applicable only if a value for nfolds is specified and fold_column is not selected) Select the cross-validation fold assignment scheme. The available options are Random or Modulo.
fold_column: (GLM, GBM, DL, DRF, K-Means) Select the column that contains the cross-validation fold index assignment per observation.
offset_column: (GLM, DRF, GBM) Select a column to use as the offset.
Note: Offsets are per-row “bias values” that are used during model training. For Gaussian distributions, they can be seen as simple corrections to the response (y) column. Instead of learning to predict the response (y-row), the model learns to predict the (row) offset of the response column. For other distributions, the offset corrections are applied in the linearized space before applying the inverse link function to get the actual response values. For more information, refer to the following link.
weights_column: (GLM, DL, DRF, GBM) Select a column to use for the observation weights. The specified
weights_columnmust be included in the specifiedtraining_frame. Python only: To use a weights column when passing an H2OFrame toxinstead of a list of column names, the specifiedtraining_framemust contain the specifiedweights_column.Note: Weights are per-row observation weights and do not increase the size of the data frame. This is typically the number of times a row is repeated, but non-integer values are supported as well. During training, rows with higher weights matter more, due to the larger loss function pre-factor.
loss: (DL) Select the loss function. For DL, the options are Automatic, Quadratic, CrossEntropy, Huber, or Absolute and the default value is Automatic. Absolute, Quadratic, and Huber are applicable for regression or classification, while CrossEntropy is only applicable for classification. Huber can improve for regression problems with outliers.
checkpoint: (DL, DRF, GBM) Enter a model key associated with a previously-trained model. Use this option to build a new model as a continuation of a previously-generated model.
use_all_factor_levels: (DL, PCA) Check this checkbox to use all factor levels in the possible set of predictors; if you enable this option, sufficient regularization is required. By default, the first factor level is skipped. For Deep Learning models, this option is useful for determining variable importances and is automatically enabled if the autoencoder is selected.
train_samples_per_iteration: (DL) Specify the number of global training samples per MapReduce iteration. To specify one epoch, enter 0. To specify all available data (e.g., replicated training data), enter -1. To use the automatic values, enter -2.
adaptive_rate: (DL) Check this checkbox to enable the adaptive learning rate (ADADELTA). This option is selected by default. If this option is enabled, the following parameters are ignored:
rate,rate_decay,rate_annealing,momentum_start,momentum_ramp,momentum_stable, andnesterov_accelerated_gradient.input_dropout_ratio: (DL) Specify the input layer dropout ratio to improve generalization. Suggested values are 0.1 or 0.2. The range is >= 0 to <1.
l1: (DL) Specify the L1 regularization to add stability and improve generalization; sets the value of many weights to 0.
l2: (DL) Specify the L2 regularization to add stability and improve generalization; sets the value of many weights to smaller values.
balance_classes: (GBM, DL) Oversample the minority classes to balance the class distribution. This option is not selected by default and can increase the data frame size. This option is only applicable for classification. Majority classes can be undersampled to satisfy the Max_after_balance_size parameter.
Note:
balance_classesbalances over just the target, not over all classes in the training frame.max_confusion_matrix_size: (DRF, DL, Naïve Bayes, GBM, GLM) Specify the maximum size (in number of classes) for confusion matrices to be printed in the Logs.
max_hit_ratio_k: (DRF, DL, Naïve Bayes, GBM, GLM) Specify the maximum number (top K) of predictions to use for hit ratio computation. Applicable to multinomial only. To disable, enter 0.
r2_stopping: (GBM, DRF) Specify a threshold for the coefficient of determination (r^2) metric value. When this threshold is met or exceeded, H2O stops making trees.
build_tree_one_node: (DRF, GBM) To run on a single node, check this checkbox. This is suitable for small datasets as there is no network overhead but fewer CPUs are used. The default setting is disabled.
rate: (DL) Specify the learning rate. Higher rates result in less stable models and lower rates result in slower convergence. Not applicable if adaptive_rate is enabled.
rate_annealing: (DL) Specify the learning rate annealing. The formula is rate/(1+rate_annealing value * samples). Not applicable if adaptive_rate is enabled.
momentum_start: (DL) Specify the initial momentum at the beginning of training. A suggested value is 0.5. Not applicable if adaptive_rate is enabled.
momentum_ramp: (DL) Specify the number of training samples for increasing the momentum. Not applicable if adaptive_rate is enabled.
momentum_stable: (DL) Specify the final momentum value reached after the momentum_ramp training samples. Not applicable if adaptive_rate is enabled.
nesterov_accelerated_gradient: (DL) Check this checkbox to use the Nesterov accelerated gradient. This option is recommended and selected by default. Not applicable is adaptive_rate is enabled.
hidden_dropout_ratios: (DL) Specify the hidden layer dropout ratios to improve generalization. Specify one value per hidden layer, each value between 0 and 1 (exclusive). There is no default value. This option is applicable only if TanhwithDropout, RectifierwithDropout, or MaxoutWithDropout is selected from the Activation drop-down list.
tweedie_power: (DL, GBM) (Only applicable if Tweedie is selected for Family) Specify the Tweedie power. The range is from 1 to 2. For a normal distribution, enter
0. For Poisson distribution, enter1. For a gamma distribution, enter2. For a compound Poisson-gamma distribution, enter a value greater than 1 but less than 2. For more information, refer to Tweedie distribution.score_interval: (DL) Specify the shortest time interval (in seconds) to wait between model scoring.
score_training_samples: (DL) Specify the number of training set samples for scoring. To use all training samples, enter 0.
score_validation_samples: (DL) (Requires selection from the validation_frame drop-down list) This option is applicable to classification only. Specify the number of validation set samples for scoring. To use all validation set samples, enter 0.
score_duty_cycle: (DL) Specify the maximum duty cycle fraction for scoring. A lower value results in more training and a higher value results in more scoring. The value must be greater than 0 and less than 1.
autoencoder: (DL) Check this checkbox to enable the Deep Learning autoencoder. This option is not selected by default.
Note: This option requires a loss function other than CrossEntropy. If this option is enabled, use_all_factor_levels must be enabled.
Expert Options
keep_cross_validation_predictions: (GLM, GBM, DL, DRF, K-Means) To keep the cross-validation predictions, check this checkbox.
class_sampling_factors: (DRF, GBM, DL) Specify the per-class (in lexicographical order) over/under-sampling ratios. By default, these ratios are automatically computed during training to obtain the class balance. This option is only applicable for classification problems and when balance_classes is enabled.
overwrite_with_best_model: (DL) Check this checkbox to overwrite the final model with the best model found during training. This option is selected by default.
target_ratio_comm_to_comp: (DL) Specify the target ratio of communication overhead to computation. This option is only enabled for multi-node operation and if train_samples_per_iteration equals -2 (auto-tuning).
rho: (DL) Specify the adaptive learning rate time decay factor. This option is only applicable if adaptive_rate is enabled.
epsilon: (DL) Specify the adaptive learning rate time smoothing factor to avoid dividing by zero. This option is only applicable if adaptive_rate is enabled.
max_w2: (DL) Specify the constraint for the squared sum of the incoming weights per unit (e.g., for Rectifier).
initial_weight_distribution: (DL) Select the initial weight distribution (Uniform Adaptive, Uniform, or Normal). If Uniform Adaptive is used, the initial_weight_scale parameter is not applicable.
initial_weight_scale: (DL) Specify the initial weight scale of the distribution function for Uniform or Normal distributions. For Uniform, the values are drawn uniformly from initial weight scale. For Normal, the values are drawn from a Normal distribution with the standard deviation of the initial weight scale. If Uniform Adaptive is selected as the initial_weight_distribution, the initial_weight_scale parameter is not applicable.
classification_stop: (DL) (Applicable to discrete/categorical datasets only) Specify the stopping criterion for classification error fractions on training data. To disable this option, enter -1.
max_hit_ratio_k: (DL, GLM) (Classification only) Specify the maximum number (top K) of predictions to use for hit ratio computation (for multinomial only). To disable this option, enter 0.
regression_stop: (DL) (Applicable to real value/continuous datasets only) Specify the stopping criterion for regression error (MSE) on the training data. To disable this option, enter -1.
diagnostics: (DL) Check this checkbox to compute the variable importances for input features (using the Gedeon method). For large networks, selecting this option can reduce speed. This option is selected by default.
fast_mode: (DL) Check this checkbox to enable fast mode, a minor approximation in back-propagation. This option is selected by default.
force_load_balance: (DL) Check this checkbox to force extra load balancing to increase training speed for small datasets and use all cores. This option is selected by default.
single_node_mode: (DL) Check this checkbox to force H2O to run on a single node for fine-tuning of model parameters. This option is not selected by default.
replicate_training_data: (DL) Check this checkbox to replicate the entire training dataset on every node for faster training on small datasets. This option is not selected by default. This option is only applicable for clouds with more than one node.
shuffle_training_data: (DL) Check this checkbox to shuffle the training data. This option is recommended if the training data is replicated and the value of train_samples_per_iteration is close to the number of nodes times the number of rows. This option is not selected by default.
missing_values_handling: (DL) Select how to handle missing values (Skip or MeanImputation).
quiet_mode: (DL) Check this checkbox to display less output in the standard output. This option is not selected by default.
sparse: (DL) Check this checkbox to enable sparse data handling, which is more efficient for data with many zero values.
col_major: (DL) Check this checkbox to use a column major weight matrix for the input layer. This option can speed up forward propagation but may reduce the speed of backpropagation. This option is not selected by default.
Note: This parameter has been deprecated.
average_activation: (DL) Specify the average activation for the sparse autoencoder. If Rectifier is selected as the Activation type, this value must be positive. For Tanh, the value must be in (-1,1).
sparsity_beta: (DL) Specify the sparsity-based regularization optimization. For more information, refer to the following link.
max_categorical_features: (DL) Specify the maximum number of categorical features enforced via hashing.
reproducible: (DL) To force reproducibility on small data, check this checkbox. If this option is enabled, the model takes more time to generate, since it uses only one thread.
export_weights_and_biases: (DL) To export the neural network weights and biases as H2O frames, check this checkbox.
max_after_balance_size: (DRF, GBM, DL) Specify the maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes.
nbins_top_level: (DRF, GBM) (For numerical [real/int] columns only) Specify the maximum number of bins at the root level to use to build the histogram. This number will then be decreased by a factor of two per level.
seed: (K-Means, GBM, DL, DRF) Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations.
intercept: (GLM) To include a constant term in the model, check this checkbox. This option is selected by default.
objective_epsilon: (GLM) Specify a threshold for convergence. If the objective value is less than this threshold, the model is converged.
beta_epsilon: (GLM) Specify the beta epsilon value. If the L1 normalization of the current beta change is below this threshold, consider using convergence.
gradient_epsilon: (GLM) (For L-BFGS only) Specify a threshold for convergence. If the objective value (using the L-infinity norm) is less than this threshold, the model is converged.
prior: (GLM) Specify prior probability for y ==1. Use this parameter for logistic regression if the data has been sampled and the mean of response does not reflect reality.
max_active_predictors: (GLM) Specify the maximum number of active predictors during computation. This value is used as a stopping criterium to prevent expensive model building with many predictors.
Viewing Models
Click the Assist Me! button, then click the getModels link, or enter getModels in the cell in CS mode and press Ctrl+Enter. A list of available models displays.
To view all current models, you can also click the Model menu and click List All Models.
To inspect a model, check its checkbox then click the Inspect button, or click the Inspect button to the right of the model name.
A summary of the model’s parameters displays. To display more details, click the Show All Parameters button.
To delete a model, click the Delete button.
To generate a Plain Old Java Object (POJO) that can use the model outside of H2O, click the Download POJO button.
Note: A POJO can be run in standalone mode or it can be integrated into a platform, such as Hadoop’s Storm. To make the POJO work in your Java application, you will also need the
h2o-genmodel.jarfile (available inh2o-3/h2o-genmodel/build/libs/h2o-genmodel.jar).
Exporting and Importing Models
To export a built model:
- Click the Model menu at the top of the screen.
- Select Export Model…
- In the
exportModelcell that appears, select the model from the drop-down Model: list. - Enter a location for the exported model in the Path: entry field.
Note: If you specify a location that doesn’t exist, it will be created. For example, if you only enter
testin the Path: entry field, the model will be exported toh2o-3/test. - To overwrite any files with the same name, check the Overwrite: checkbox.
Click the Export button. A confirmation message displays when the model has been successfully exported.
To import a built model:
- Click the Model menu at the top of the screen.
- Select Import Model…
- Enter the location of the model in the Path: entry field.
Note: The file path must be complete (e.g.,
Users/h2o-user/h2o-3/exported_models). Do not rename models while importing. - To overwrite any files with the same name, check the Overwrite: checkbox.
Click the Import button. A confirmation message displays when the model has been successfully imported. To view the imported model, click the View Model button.
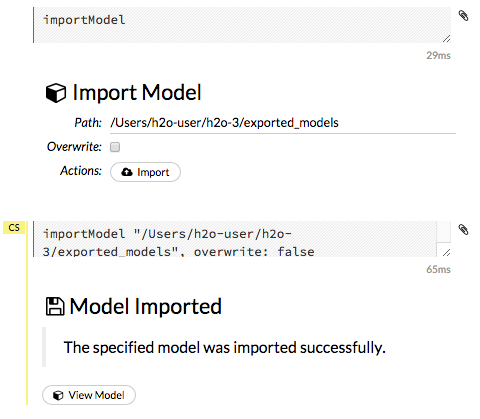
Using Grid Search
To include a parameter in a grid search in Flow, check the checkbox in the Grid? column to the right of the parameter name (highlighted in red in the image below).

- If the parameter selected for grid search is Boolean (T/F or Y/N), both values are included when the Grid? checkbox is selected.
- If the parameter selected for grid search is a list of values, the values display as checkboxes when the Grid? checkbox is selected. More than one option can be selected.
- If the parameter selected for grid search is a numerical value, use a semicolon (;) to separate each additional value.
- To view a list of all grid searches, select the Model menu, then click List All Grid Search Results, or click the Assist Me button and select getGrids.
Checkpointing Models
Some model types, such as DRF, GBM, and Deep Learning, support checkpointing. A checkpoint resumes model training so that you can iterate your model. The dataset must be the same. The following model parameters must be the same when restarting a model from a checkpoint:
| Must be the same as in checkpoint model | |||
|---|---|---|---|
drop_na20_cols |
response_column |
activation |
|
use_all_factor_levels |
adaptive_rate |
autoencoder |
|
rho |
epsilon |
sparse |
|
sparsity_beta |
col_major |
rate |
|
rate_annealing |
rate_decay |
momentum_start |
|
momentum_ramp |
momentum_stable |
nesterov_accelerated_gradient |
|
ignore_const_cols |
max_categorical_features |
nfolds |
|
distribution |
tweedie_power |
The following parameters can be modified when restarting a model from a checkpoint:
| Can be modified | |||
|---|---|---|---|
seed |
checkpoint |
epochs |
|
score_interval |
train_samples_per_iteration |
target_ratio_comm_to_comp |
|
score_duty_cycle |
score_training_samples |
score_validation_samples |
|
score_validation_sampling |
classification_stop |
regression_stop |
|
quiet_mode |
max_confusion_matrix_size |
max_hit_ratio_k |
|
diagnostics |
variable_importances |
initial_weight_distribution |
|
initial_weight_scale |
force_load_balance |
replicate_training_data |
|
shuffle_training_data |
single_node_mode |
fast_mode |
|
l1 |
l2 |
max_w2 |
|
input_dropout_ratio |
hidden_dropout_ratios |
loss |
|
overwrite_with_best_model |
missing_values_handling |
average_activation |
|
reproducible |
export_weights_and_biases |
elastic_averaging |
|
elastic_averaging_moving_rate |
elastic_averaging_regularization |
mini_batch_size |
- After building your model, copy the
model_id. To view themodel_id, click the Model menu then click List All Models. - Select the model type from the drop-down Model menu.
Note: The model type must be the same as the checkpointed model.
- Paste the copied
model_idin the checkpoint entry field. - Click the Build Model button. The model will resume training.
Interpreting Model Results
Scoring history: GBM, DL Represents the error rate of the model as it is built. Typically, the error rate will be higher at the beginning (the left side of the graph) then decrease as the model building completes and accuracy improves. Can include mean squared error (MSE) and deviance.
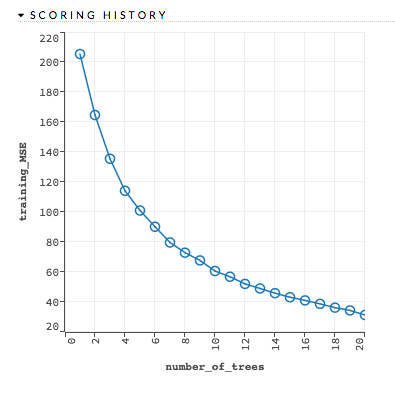
Variable importances: GBM, DL Represents the statistical significance of each variable in the data in terms of its affect on the model. Variables are listed in order of most to least importance. The percentage values represent the percentage of importance across all variables, scaled to 100%. The method of computing each variable’s importance depends on the algorithm. To view the scaled importance value of a variable, use your mouse to hover over the bar representing the variable.
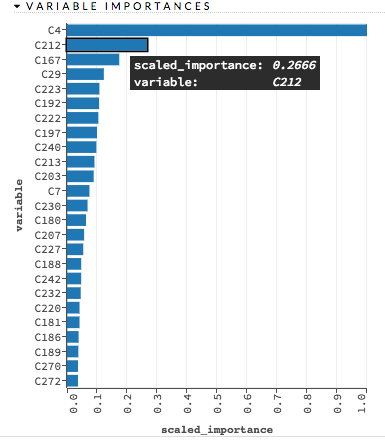
Confusion Matrix: DL Table depicting performance of algorithm in terms of false positives, false negatives, true positives, and true negatives. The actual results display in the columns and the predictions display in the rows; correct predictions are highlighted in yellow. In the example below, 0 was predicted correctly 902 times, while 8 was predicted correctly 822 times and 0 was predicted as 4 once.
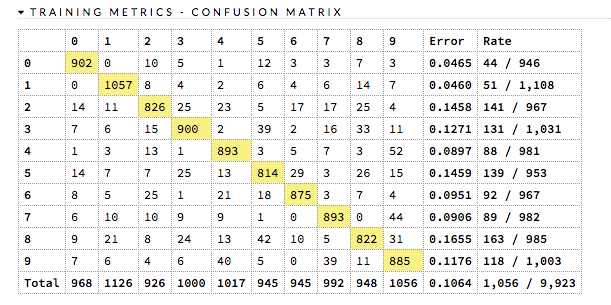
ROC Curve: DL, GLM, DRF Graph representing the ratio of true positives to false positives. To view a specific threshold, select a value from the drop-down Threshold list. To view any of the following details, select it from the drop-down Criterion list:
- Max f1
- Max f2
- Max f0point5
- Max accuracy
- Max precision
- Max absolute MCC (the threshold that maximizes the absolute Matthew’s Correlation Coefficient)
- Max min per class accuracy
The lower-left side of the graph represents less tolerance for false positives while the upper-right represents more tolerance for false positives. Ideally, a highly accurate ROC resembles the following example.
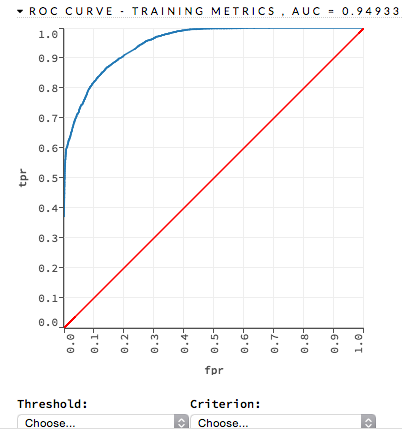
Hit Ratio: GBM, DRF, NaiveBayes, DL, GLM (Multinomial Classification only) Table representing the number of times that the prediction was correct out of the total number of predictions.
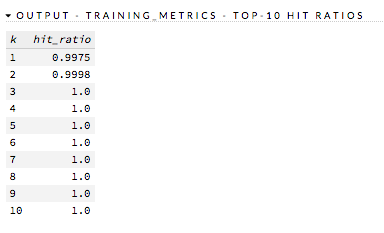
Standardized Coefficient Magnitudes GLM Bar chart representing the relationship of a specific feature to the response variable. Coefficients can be positive (orange) or negative (blue). A positive coefficient indicates a positive relationship between the feature and the response, where an increase in the feature corresponds with an increase in the response, while a negative coefficient represents a negative relationship between the feature and the response where an increase in the feature corresponds with a decrease in the response (or vice versa).
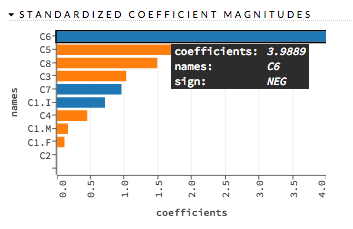
To learn how to make predictions, continue to the next section.
… Making Predictions
After creating your model, click the key link for the model, then click the Predict button. Select the model to use in the prediction from the drop-down Model: menu and the data frame to use in the prediction from the drop-down Frame: menu, then click the Predict button.
Viewing Predictions
Click the Assist Me! button, then click the getPredictions link, or enter getPredictions in the cell in CS mode and press Ctrl+Enter. A list of the stored predictions displays.
To view a prediction, click the View button to the right of the model name.
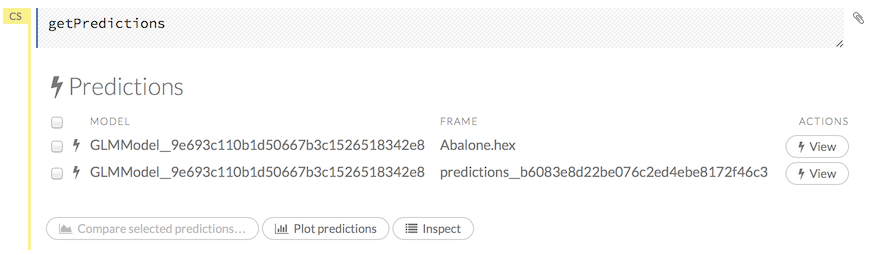
You can also view predictions by clicking the drop-down Score menu and selecting List All Predictions.
Viewing Frames
To view a specific frame, click the “Key” link for the specified frame, or enter getFrameSummary "FrameName" in a cell in CS mode (where FrameName is the name of a frame, such as allyears2k.hex).
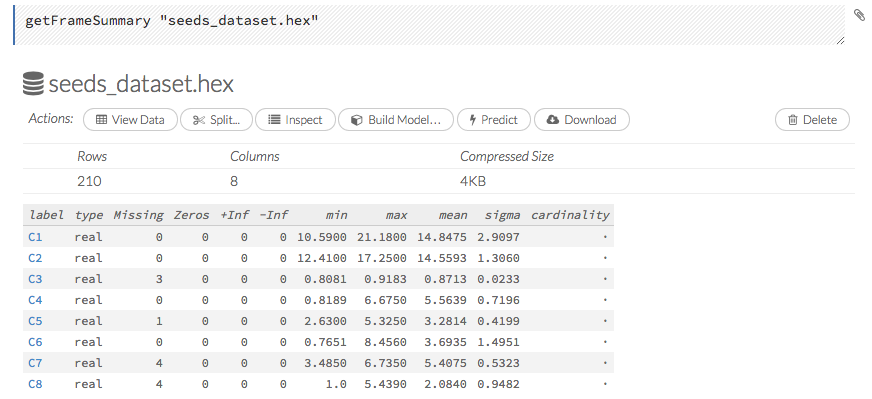
From the getFrameSummary cell, you can:
- view a truncated list of the rows in the data frame by clicking the View Data button
- split the dataset by clicking the Split… button
- view the columns, data, and factors in more detail or plot a graph by clicking the Inspect button
- create a model by clicking the Build Model button
- make a prediction based on the data by clicking the Predict button
- download the data as a .csv file by clicking the Download button
- view the characteristics or domain of a specific column by clicking the Summary link
When you view a frame, you can “drill-down” to the necessary level of detail (such as a specific column or row) using the Inspect button or by clicking the links. The following screenshot displays the results of clicking the Inspect button for a frame.
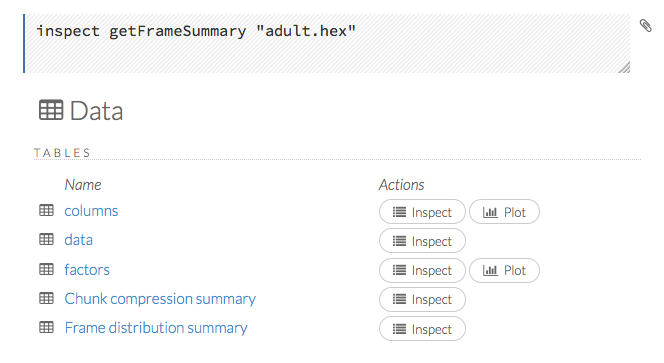
This screenshot displays the results of clicking the columns link.
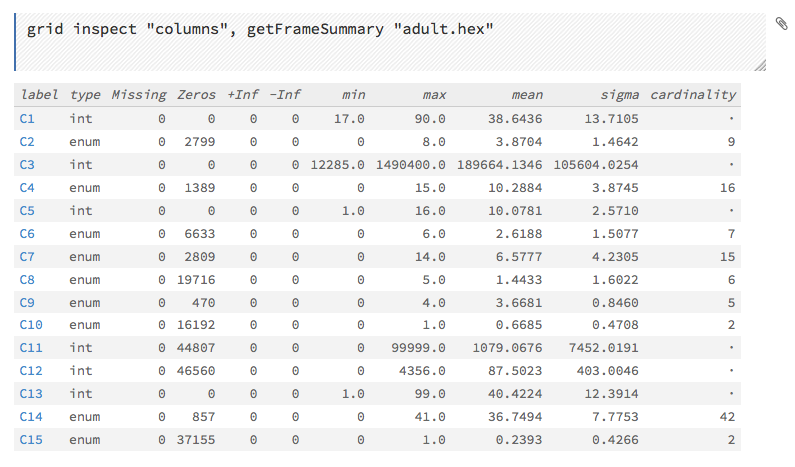
To view all frames, click the Assist Me! button, then click the getFrames link, or enter getFrames in the cell in CS mode and press Ctrl+Enter. You can also view all current frames by clicking the drop-down Data menu and selecting List All Frames.
A list of the current frames in H2O displays that includes the following information for each frame:
- Link to the frame (the “key”)
- Number of rows and columns
- Size
For parsed data, the following information displays:
- Link to the .hex file
The Build Model, Predict, and Inspect buttons
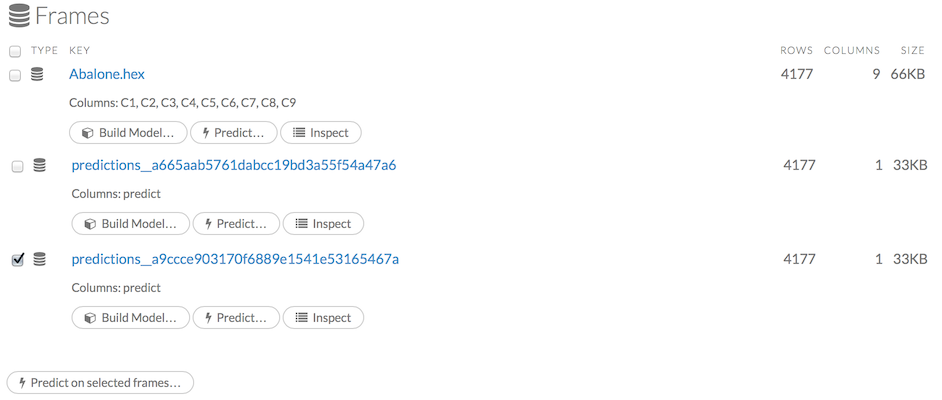
To make a prediction, check the checkboxes for the frames you want to use to make the prediction, then click the Predict on Selected Frames button.
Splitting Frames
Datasets can be split within Flow for use in model training and testing.
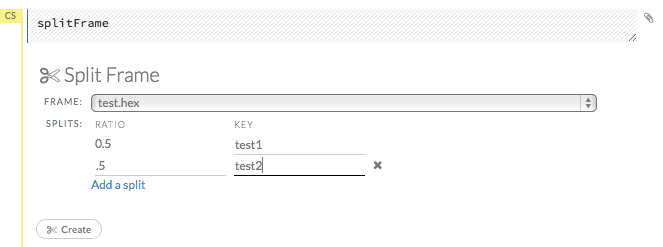
To split a frame, click the Assist Me button, then click splitFrame.
Note: You can also click the drop-down Data menu and select Split Frame….
- From the drop-down Frame: list, select the frame to split.
In the second Ratio entry field, specify the fractional value to determine the split. The first Ratio field is automatically calculated based on the values entered in the second Ratio field.
Note: Only fractional values between 0 and 1 are supported (for example, enter
.5to split the frame in half). The total sum of the ratio values must equal one. H2O automatically adjusts the ratio values to equal one; if unsupported values are entered, an error displays.- In the Key entry field, specify a name for the new frame.
- (Optional) To add another split, click the Add a split link. To remove a split, click the
Xto the right of the Key entry field. - Click the Create button.
Creating Frames
To create a frame with a large amount of random data (for example, to use for testing), click the drop-down Admin menu, then select Create Synthetic Frame. Customize the frame as needed, then click the Create button to create the frame.
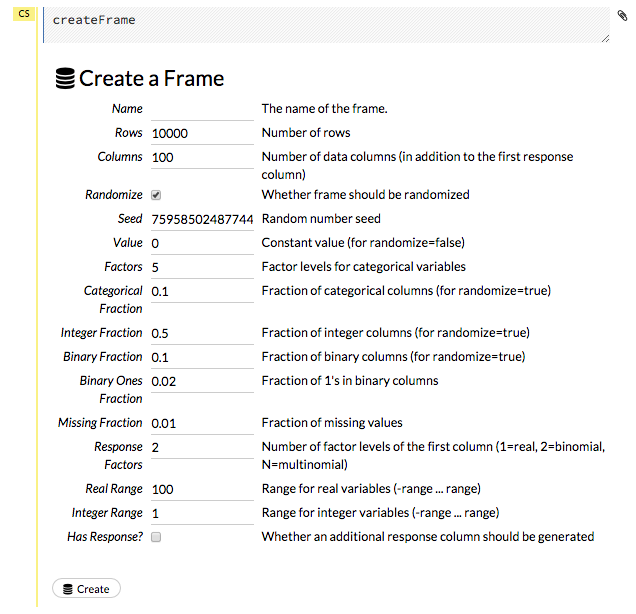
Plotting Frames
To create a plot from a frame, click the Inspect button, then click the Plot button.
Select the type of plot (point, path, or rect) from the drop-down Type menu, then select the x-axis and y-axis from the following options:
- label
- type
- missing
- zeros
- +Inf
- -Inf
- min
- max
- mean
- sigma
- cardinality
Select one of the above options from the drop-down Color menu to display the specified data in color, then click the Plot button to plot the data.
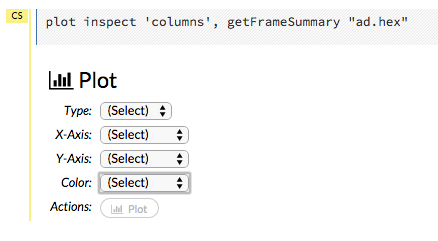
Note: Because H2O stores enums internally as numeric then maps the integers to an array of strings, any
min,max, ormeanvalues for categorical columns are not meaningful and should be ignored. Displays for categorical data will be modified in a future version of H2O.
… Using Flows
You can use and modify flows in a variety of ways:
- Clips allow you to save single cells
- Outlines display summaries of your workflow
- Flows can be saved, duplicated, loaded, or downloaded
Using Clips
Clips enable you to save cells containing your workflow for later reuse. To save a cell as a clip, click the paperclip icon to the right of the cell (highlighted in the red box in the following screenshot).

To use a clip in a workflow, click the “Clips” tab in the sidebar on the right.
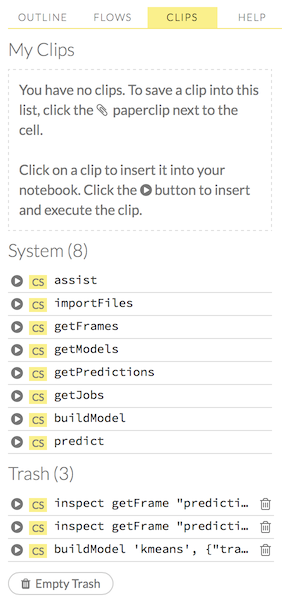
All saved clips, including the default system clips (such as assist, importFiles, and predict), are listed. Clips you have created are listed under the “My Clips” heading. To select a clip to insert, click the circular button to the left of the clip name. To delete a clip, click the trashcan icon to right of the clip name.
NOTE: The default clips listed under “System” cannot be deleted.
Deleted clips are stored in the trash. To permanently delete all clips in the trash, click the Empty Trash button.
NOTE: Saved data, including flows and clips, are persistent as long as the same IP address is used for the cluster. If a new IP is used, previously saved flows and clips are not available.
Viewing Outlines
The Outline tab in the sidebar displays a brief summary of the cells currently used in your flow; essentially, a command history.
- To jump to a specific cell, click the cell description.
To delete a cell, select it and press the X key on your keyboard.
Saving Flows
- Finding Saved Flows on your Disk
- Saving Flows on a Hadoop cluster
- Copying Flows
- Downloading Flows
- Loading Flows
You can save your flow for later reuse. To save your flow as a notebook, click the “Save” button (the first button in the row of buttons below the flow name), or click the drop-down “Flow” menu and select “Save Flow.” To enter a custom name for the flow, click the default flow name (“Untitled Flow”) and type the desired flow name. A pencil icon indicates where to enter the desired name.

To confirm the name, click the checkmark to the right of the name field.
To reuse a saved flow, click the “Flows” tab in the sidebar, then click the flow name. To delete a saved flow, click the trashcan icon to the right of the flow name.
Finding Saved Flows on your Disk
By default, flows are saved to the h2oflows directory underneath your home directory. The directory where flows are saved is printed to stdout:
03-20 14:54:20.945 172.16.2.39:54323 95667 main INFO: Flow dir: '/Users/<UserName>/h2oflows'
To back up saved flows, copy this directory to your preferred backup location.
To specify a different location for saved flows, use the command-line argument -flow_dir when launching H2O:
java -jar h2o.jar -flow_dir /<New>/<Location>/<For>/<Saved>/<Flows>
where /<New>/<Location>/<For>/<Saved>/<Flows> represents the specified location. If the directory does not exist, it will be created the first time you save a flow.
Saving Flows on a Hadoop cluster
If you are running H2O Flow on a Hadoop cluster, H2O will try to find the HDFS home directory to use as the default directory for flows. If the HDFS home directory is not found, flows cannot be saved unless a directory is specified while launching using -flow_dir:
hadoop jar h2odriver.jar -nodes 1 -mapperXmx 6g -output hdfsOutputDirName -flow_dir hdfs:///<Saved>/<Flows>/<Location>
The location specified in flow_dir may be either an hdfs or regular filesystem directory. If the directory does not exist, it will be created the first time you save a flow.
Copying Flows
To create a copy of the current flow, select the Flow menu, then click Make a Copy. The name of the current flow changes to Copy of <FlowName> (where <FlowName> is the name of the flow). You can save the duplicated flow using this name by clicking Flow > Save Flow, or rename it before saving.
Downloading Flows
After saving a flow as a notebook, click the Flow menu, then select Download this Flow. A new window opens and the saved flow is downloaded to the default downloads folder on your computer. The file is exported as <filename>.flow, where <filename> is the name specified when the flow was saved.
Caution: You must have an active internet connection to download flows.
Loading Flows
To load a saved flow, click the Flows tab in the sidebar at the right. In the pop-up confirmation window that appears, select Load Notebook, or click Cancel to return to the current flow.
After clicking Load Notebook, the saved flow is loaded.
To load an exported flow, click the Flow menu and select Open Flow…. In the pop-up window that appears, click the Choose File button and select the exported flow, then click the Open button.
Notes:
- Only exported flows using the default .flow filetype are supported. Other filetypes will not open.
- If the current notebook has the same name as the selected file, a pop-up confirmation appears to confirm that the current notebook should be overwritten.
…Troubleshooting Flow
- Viewing Cluster Status
- Viewing CPU Status (Water Meter)
- Viewing Logs
- Downloading Logs
- Viewing Stack Trace Information
- Viewing Network Test Results
- Accessing the Profiler
- Viewing the Timeline
- Reporting Issues
- Requesting Help
- Shutting Down H2O
To troubleshoot issues in Flow, use the Admin menu. The Admin menu allows you to check the status of the cluster, view a timeline of events, and view or download logs for issue analysis.
NOTE: To view the current H2O Flow version, click the Help menu, then click About.
Viewing Cluster Status
Click the Admin menu, then select Cluster Status. A summary of the status of the cluster (also known as a cloud) displays, which includes the same information:
- Cluster health
- Whether all nodes can communicate (consensus)
Whether new nodes can join (locked/unlocked)
Note: After you submit a job to H2O, the cluster does not accept new nodes.
- H2O version
- Number of used and available nodes
When the cluster was created
The following information displays for each node:
- IP address (name)
- Time of last ping
- Number of cores
- Load
- Amount of data (used/total)
- Percentage of cached data
- GC (free/total/max)
- Amount of disk space in GB (free/max)
- Percentage of free disk space
To view more information, click the Show Advanced button.
Viewing CPU Status (Water Meter)
To view the current CPU usage, click the Admin menu, then click Water Meter (CPU Meter). A new window opens, displaying the current CPU use statistics.
Viewing Logs
To view the logs for troubleshooting, click the Admin menu, then click Inspect Log.
To view the logs for a specific node, select it from the drop-down Select Node menu.
Downloading Logs
To download the logs for further analysis, click the Admin menu, then click Download Log. A new window opens and the logs download to your default download folder. You can close the new window after downloading the logs. Send the logs to h2ostream or file a JIRA ticket for issue resolution.
Viewing Stack Trace Information
To view the stack trace information, click the Admin menu, then click Stack Trace.
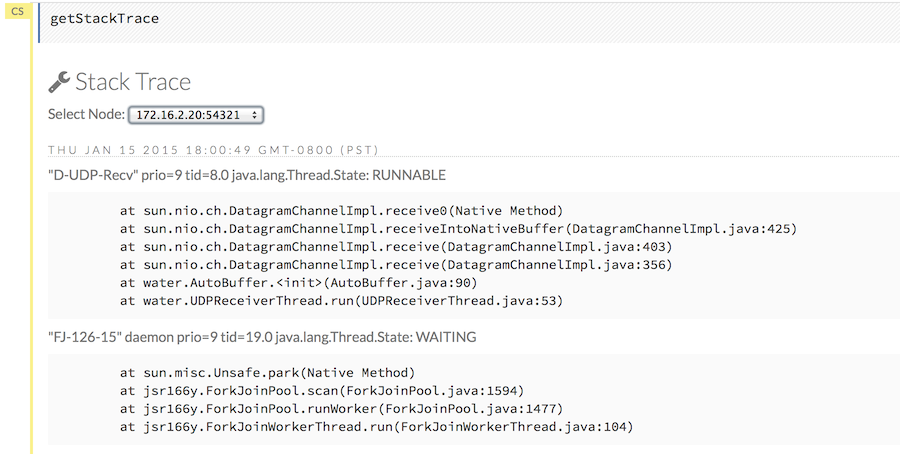
To view the stack trace information for a specific node, select it from the drop-down Select Node menu.
Viewing Network Test Results
To view network test results, click the Admin menu, then click Network Test.
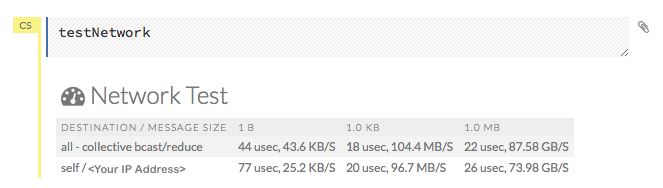
Accessing the Profiler
The Profiler looks across the cluster to see where the same stack trace occurs, and can be helpful for identifying activity on the current CPU. To view the profiler, click the Admin menu, then click Profiler.
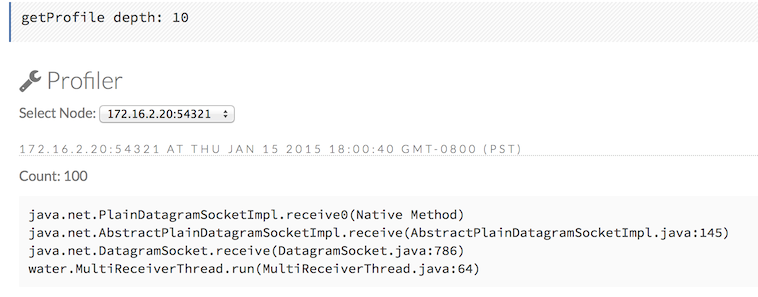
To view the profiler information for a specific node, select it from the drop-down Select Node menu.
Viewing the Timeline
To view a timeline of events in Flow, click the Admin menu, then click Timeline. The following information displays for each event:
- Time of occurrence (HH:MM:SS:MS)
- Number of nanoseconds for duration
- Originator of event (“who”)
- I/O type
- Event type
Number of bytes sent & received
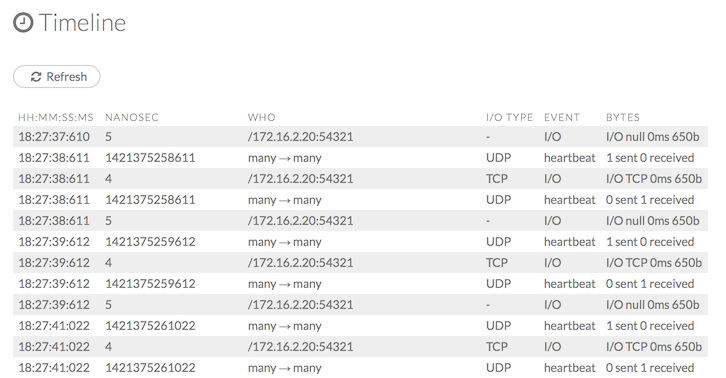
To obtain the most recent information, click the Refresh button.
Reporting Issues
If you experience an error with Flow, you can submit a JIRA ticket to notify our team.
- First, click the Admin menu, then click Download Logs. This will download a file contains information that will help our developers identify the cause of the issue.
- Click the Help menu, then click Report an issue. This will open our JIRA page where you can file your ticket.
- Click the Create button at the top of the JIRA page.
- Attach the log file from the first step, write a description of the error you experienced, then click the Create button at the bottom of the page. Our team will work to resolve the issue and you can track the progress of your ticket in JIRA.
Requesting Help
If you have a Google account, you can submit a request for assistance with H2O on our Google Groups page, H2Ostream.
To access H2Ostream from Flow:
- Click the Help menu.
- Click Forum/Ask a question.
- Click the red New topic button.
- Enter your question and click the red Post button. If you are requesting assistance for an error you experienced, be sure to include your logs.
You can also email your question to h2ostream@googlegroups.com.
Shutting Down H2O
To shut down H2O, click the Admin menu, then click Shut Down. A Shut down complete message displays in the upper right when the cluster has been shut down.
Data Science Algorithms
This document describes how to define the models and how to interpret the model, as well the algorithm itself, and provides an FAQ.
Commonalities
Quantiles
Note: The quantile results in Flow are computed lazily on-demand and cached. It is a fast approximation (max - min / 1024) that is very accurate for most use cases.
If the distribution is skewed, the quantile results may not be as accurate as the results obtained using h2o.quantile in R or H2OFrame.quantile in Python.
K-Means
Introduction
K-Means falls in the general category of clustering algorithms.
Defining a K-Means Model
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: (Optional) Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
k*: Specify the number of clusters.
user_points: Specify a vector of initial cluster centers. The user-specified points must have the same number of columns as the training observations. The number of rows must equal the number of clusters.
max_iterations: Specify the maximum number of training iterations. The range is 0 to 1e6.
init: Select the initialization mode. The options are Random, Furthest, PlusPlus, or User. Note: If PlusPlus is selected, the initial Y matrix is chosen by the final cluster centers from the K-Means PlusPlus algorithm.
fold_assignment: (Applicable only if a value for nfolds is specified and fold_column is not selected) Select the cross-validation fold assignment scheme. The available options are AUTO (which is Random), Random, or Modulo.
fold_column: Select the column that contains the cross-validation fold index assignment per observation.
score_each_iteration: (Optional) Check this checkbox to score during each iteration of the model training.
standardize: To standardize the numeric columns to have mean of zero and unit variance, check this checkbox. Standardization is highly recommended; if you do not use standardization, the results can include components that are dominated by variables that appear to have larger variances relative to other attributes as a matter of scale, rather than true contribution. This option is selected by default.
Note: If standardization is enabled, each column of numeric data is centered and scaled so that its mean is zero and its standard deviation is one before the algorithm is used. At the end of the process, the cluster centers on both the standardized scale (
centers_std) and the de-standardized scale (centers) are displayed. To de-standardize the centers, the algorithm multiplies by the original standard deviation of the corresponding column and adds the original mean. Enabling standardization is mathematically equivalent to usingh2o.scalein R withcenter= TRUE andscale= TRUE on the numeric columns. Therefore, there will be no discernible difference if standardization is enabled or not for K-Means, since H2O calculates unstandardized centroids.keep_cross_validation_predictions: To keep the cross-validation predictions, check this checkbox.
seed: Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations.
Interpreting a K-Means Model
By default, the following output displays:
- A graph of the scoring history (number of iterations vs. average within the cluster’s sum of squares)
- Output (model category, validation metrics if applicable, and centers std)
- Model Summary (number of clusters, number of categorical columns, number of iterations, avg. within sum of squares, avg. sum of squares, avg. between the sum of squares)
- Scoring history (number of iterations, avg. change of standardized centroids, avg. within cluster sum of squares)
- Training metrics (model name, checksum name, frame name, frame checksum name, description if applicable, model category, duration in ms, scoring time, predictions, MSE, avg. within sum of squares, avg. between sum of squares)
- Centroid statistics (centroid number, size, within sum of squares)
- Cluster means (centroid number, column)
K-Means randomly chooses starting points and converges to a local minimum of centroids. The number of clusters is arbitrary, and should be thought of as a tuning parameter. The output is a matrix of the cluster assignments and the coordinates of the cluster centers in terms of the originally chosen attributes. Your cluster centers may differ slightly from run to run as this problem is Non-deterministic Polynomial-time (NP)-hard.
FAQ
How does the algorithm handle missing values during training?
Missing values are automatically imputed by the column mean. K-means also handles missing values by assuming that missing feature distance contributions are equal to the average of all other distance term contributions.
How does the algorithm handle missing values during testing?
Missing values are automatically imputed by the column mean of the training data.
Does it matter if the data is sorted?
No.
Should data be shuffled before training?
No.
What if there are a large number of columns?
K-Means suffers from the curse of dimensionality: all points are roughly at the same distance from each other in high dimensions, making the algorithm less and less useful.
What if there are a large number of categorical factor levels?
This can be problematic, as categoricals are one-hot encoded on the fly, which can lead to the same problem as datasets with a large number of columns.
K-Means Algorithm
The number of clusters \(K\) is user-defined and is determined a priori.
Choose \(K\) initial cluster centers \(m_{k}\) according to one of the following:
Randomization: Choose \(K\) clusters from the set of \(N\) observations at random so that each observation has an equal chance of being chosen.
Plus Plus
a. Choose one center \(m_{1}\) at random.
Calculate the difference between \(m_{1}\) and each of the remaining \(N-1\) observations \(x_{i}\). \(d(x_{i}, m_{1}) = ||(x_{i}-m_{1})||^2\)
Let \(P(i)\) be the probability of choosing \(x_{i}\) as \(m_{2}\). Weight \(P(i)\) by \(d(x_{i}, m_{1})\) so that those \(x_{i}\) furthest from \(m_{2}\) have a higher probability of being selected than those \(x_{i}\) close to \(m_{1}\).
Choose the next center \(m_{2}\) by drawing at random according to the weighted probability distribution.
Repeat until \(K\) centers have been chosen.
Furthest
a. Choose one center \(m_{1}\) at random.
Calculate the difference between \(m_{1}\) and each of the remaining \(N-1\) observations \(x_{i}\). \(d(x_{i}, m_{1}) = ||(x_{i}-m_{1})||^2\)
Choose \(m_{2}\) to be the \(x_{i}\) that maximizes \(d(x_{i}, m_{1})\).
Repeat until \(K\) centers have been chosen.
Once \(K\) initial centers have been chosen calculate the difference between each observation \(x_{i}\) and each of the centers \(m_{1},...,m_{K}\), where difference is the squared Euclidean distance taken over \(p\) parameters.
\(d(x_{i}, m_{k})=\) \(\sum_{j=1}^{p}(x_{ij}-m_{k})^2=\) \(\lVert(x_{i}-m_{k})\rVert^2\)
Assign \(x_{i}\) to the cluster \(k\) defined by \(m_{k}\) that minimizes \(d(x_{i}, m_{k})\)
When all observations \(x_{i}\) are assigned to a cluster calculate the mean of the points in the cluster.
\(\bar{x}(k)=\lbrace\bar{x_{i1}},…\bar{x_{ip}}\rbrace\)
Set the \(\bar{x}(k)\) as the new cluster centers \(m_{k}\). Repeat steps 2 through 5 until the specified number of max iterations is reached or cluster assignments of the \(x_{i}\) are stable.
References
Xiong, Hui, Junjie Wu, and Jian Chen. “K-means Clustering Versus Validation Measures: A Data- distribution Perspective.” Systems, Man, and Cybernetics, Part B: Cybernetics, IEEE Transactions on 39.2 (2009): 318-331.
GLM
Introduction
Generalized Linear Models (GLM) estimate regression models for outcomes following exponential distributions. In addition to the Gaussian (i.e. normal) distribution, these include Poisson, binomial, and gamma distributions. Each serves a different purpose, and depending on distribution and link function choice, can be used either for prediction or classification.
The GLM suite includes:
- Gaussian regression
- Poisson regression
- Binomial regression (classification)
- Multinomial classification
- Gamma regression
Defining a GLM Model
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
nfolds: Specify the number of folds for cross-validation.
Note: Lambda search is not supported when cross-validation is enabled.
response_column: (Required) Select the column to use as the independent variable.
- For a regression model, this column must be numeric (Real or Int).
- For a classification model, this column must be categorical (Enum or String). If the family is Binomial, the dataset cannot contain more than two levels.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
family: Select the model type.
- If the family is gaussian, the data must be numeric (Real or Int).
- If the family is binomial, the data must be categorical 2 levels/classes or binary (Enum or Int).
- If the family is multinomial, the data can be categorical with more than two levels/classes (Enum).
- If the family is poisson, the data must be numeric and non-negative (Int).
- If the family is gamma, the data must be numeric and continuous and positive (Real or Int).
- If the family is tweedie, the data must be numeric and continuous (Real) and non-negative.
tweedie_variance_power: (Only applicable if Tweedie is selected for Family) Specify the Tweedie variance power.
tweedie_link_power: (Only applicable if Tweedie is selected for Family) Specify the Tweedie link power.
solver: Select the solver to use (AUTO, IRLSM, L_BFGS, COORDINATE_DESCENT_NAIVE, or COORDINATE_DESCENT). IRLSM is fast on on problems with a small number of predictors and for lambda-search with L1 penalty, while L_BFGS scales better for datasets with many columns. COORDINATE_DESCENT is IRLSM with the covariance updates version of cyclical coordinate descent in the innermost loop. COORDINATE_DESCENT_NAIVE is IRLSM with the naive updates version of cyclical coordinate descent in the innermost loop. COORDINATE_DESCENT_NAIVE and COORDINATE_DESCENT are currently experimental.
alpha: Specify the regularization distribution between L2 and L2.
lambda: Specify the regularization strength.
lambda_search: Check this checkbox to enable lambda search, starting with lambda max. The given lambda is then interpreted as lambda min.
Note: Lambda search is not supported when cross-validation is enabled.
nlambdas: (Applicable only if lambda_search is enabled) Specify the number of lambdas to use in the search. The default is 100.
standardize: To standardize the numeric columns to have a mean of zero and unit variance, check this checkbox. Standardization is highly recommended; if you do not use standardization, the results can include components that are dominated by variables that appear to have larger variances relative to other attributes as a matter of scale, rather than true contribution. This option is selected by default.
remove_collinear_columns: Automatically remove collinear columns during model-building. Collinear columns will be dropped from the model and will have 0 coefficient in the returned model. Can only be set if there is no regularization (lambda=0)
compute_p_values: Request computation of p-values. Only applicable with no penalty (lambda = 0 and no beta constraints). Setting remove_collinear_columns is recommended. H2O will return an error if p-values are requested and there are collinear columns and remove_collinear_columns flag is not set.
non-negative: To force coefficients to have non-negative values, check this checkbox.
beta_constraints: To use beta constraints, select a dataset from the drop-down menu. The selected frame is used to constraint the coefficient vector to provide upper and lower bounds. The dataset must contain a names column with valid coefficient names.
fold_assignment: (Applicable only if a value for nfolds is specified and fold_column is not selected) Select the cross-validation fold assignment scheme. The available options are AUTO (which is Random), Random, or Modulo.
fold_column: Select the column that contains the cross-validation fold index assignment per observation.
score_each_iteration: (Optional) Check this checkbox to score during each iteration of the model training.
offset_column: Select a column to use as the offset; the value cannot be the same as the value for the
weights_column.Note: Offsets are per-row “bias values” that are used during model training. For Gaussian distributions, they can be seen as simple corrections to the response (y) column. Instead of learning to predict the response (y-row), the model learns to predict the (row) offset of the response column. For other distributions, the offset corrections are applied in the linearized space before applying the inverse link function to get the actual response values. For more information, refer to the following link.
weights_column: Select a column to use for the observation weights, which are used for bias correction. The specified
weights_columnmust be included in the specifiedtraining_frame. Python only: To use a weights column when passing an H2OFrame toxinstead of a list of column names, the specifiedtraining_framemust contain the specifiedweights_column.Note: Weights are per-row observation weights and do not increase the size of the data frame. This is typically the number of times a row is repeated, but non-integer values are supported as well. During training, rows with higher weights matter more, due to the larger loss function pre-factor.
max_iterations: Specify the number of training iterations.
link: Select a link function (Identity, Family_Default, Logit, Log, Inverse, or Tweedie).
- If the family is Gaussian, Identity, Log, and Inverse are supported.
- If the family is Binomial, Logit is supported.
- If the family is Poisson, Log and Identity are supported.
- If the family is Gamma, Inverse, Log, and Identity are supported.
- If the family is Tweedie, only Tweedie is supported.
- If the family is Gaussian, Identity, Log, and Inverse are supported.
max_confusion_matrix_size: Specify the maximum size (number of classes) for the confusion matrices printed in the logs.
max_hit_ratio_k: (Applicable for classification only) Specify the maximum number (top K) of predictions to use for hit ratio computation. Applicable to multi-class only. To disable, enter
0.keep_cross_validation_predictions: To keep the cross-validation predictions, check this checkbox.
intercept: To include a constant term in the model, check this checkbox. This option is selected by default.
objective_epsilon: Specify a threshold for convergence. If the objective value is less than this threshold, the model is converged.
beta_epsilon: Specify the beta epsilon value. If the L1 normalization of the current beta change is below this threshold, consider using convergence.
gradient_epsilon: (For L-BFGS only) Specify a threshold for convergence. If the objective value (using the L-infinity norm) is less than this threshold, the model is converged.
prior: Specify prior probability for p(y==1). Use this parameter for logistic regression if the data has been sampled and the mean of response does not reflect reality. Note: this is simple method affecting only the intercept, you may want to use weights and offset for better fit.
lambda_min_ratio: Specify the minimum lambda to use for lambda search (specified as a ratio of lambda_max).
max_active_predictors: Specify the maximum number of active predictors during computation. This value is used as a stopping criterium to prevent expensive model building with many predictors.
Interpreting a GLM Model
By default, the following output displays:
- A graph of the normalized coefficient magnitudes
- Output (model category, model summary, scoring history, training metrics, validation metrics, best lambda, threshold, residual deviance, null deviance, residual degrees of freedom, null degrees of freedom, AIC, AUC, binomial, rank)
- Coefficients
- Coefficient magnitudes
FAQ
How does the algorithm handle missing values during training?
Depending on the selected missing value handling policy, they are either imputed mean or the whole row is skipped.
The default behavior is mean imputation. Note that categorical variables are imputed by adding extra “missing” level.
Optionally, glm can skip all rows with any missing values.How does the algorithm handle missing values during testing? Same as during training. If the missing value handling is set to skip and we are generating predictions, skipped rows will have Na (missing) prediction.
What happens if the response has missing values?
The rows with missing response are ignored during model training and validation.
What happens during prediction if the new sample has categorical levels not seen in training? The value will be filled with either special missing level (if trained with missing values and missing_value_handling was set to MeanImputation) or 0.
Does it matter if the data is sorted?
No.
Should data be shuffled before training?
No.
How does the algorithm handle highly imbalanced data in a response column?
GLM does not require special handling for imbalanced data.
What if there are a large number of columns?
IRLS will get quadratically slower with the number of columns. Try L-BFGS for datasets with more than 5-10 thousand columns.
What if there are a large number of categorical factor levels?
GLM internally one-hot encodes the categorical factor levels; the same limitations as with a high column count will apply.
When building the model, does GLM use all features or a selection of the best features?
Typically, GLM picks the best predictors, especially if lasso is used (
alpha = 1). By default, the GLM model includes an L1 penalty and will pick only the most predictive predictors.When running GLM, is it better to create a cluster that uses many smaller nodes or fewer larger nodes?
A rough heuristic would be:
nodes ~=MN^2/(p1e8)
where M is the number of observations, N is the number of columns (categorical columns count as a single column in this case), and p is the number of CPU cores per node.
For example, a dataset with 250 columns and 1M rows would optimally use about 20 nodes with 32 cores each (following the formula 250^21000000/(321e8) = 19.5 ~= 20).
- How is variable importance calculated for GLM?
For GLM, the variable importance represents the coefficient magnitudes.
GLM Algorithm
Following the definitive text by P. McCullagh and J.A. Nelder (1989) on the generalization of linear models to non-linear distributions of the response variable Y, H2O fits GLM models based on the maximum likelihood estimation via iteratively reweighed least squares.
Let \(y_{1},…,y_{n}\) be n observations of the independent, random response variable \(Y_{i}\).
Assume that the observations are distributed according to a function from the exponential family and have a probability density function of the form:
\(f(y_{i})=exp[\frac{y_{i}\theta_{i} - b(\theta_{i})}{a_{i}(\phi)} + c(y_{i}; \phi)]\) where \(\theta\) and \(\phi\) are location and scale parameters, and \(\: a_{i}(\phi), \:b_{i}(\theta_{i}),\: c_{i}(y_{i}; \phi)\) are known functions.
\(a_{i}\) is of the form \(\:a_{i}=\frac{\phi}{p_{i}}; p_{i}\) is a known prior weight.
When \(Y\) has a pdf from the exponential family:
\(E(Y_{i})=\mu_{i}=b^{\prime}\) \(var(Y_{i})=\sigma_{i}^2=b^{\prime\prime}(\theta_{i})a_{i}(\phi)\)
Let \(g(\mu_{i})=\eta_{i}\) be a monotonic, differentiable transformation of the expected value of \(y_{i}\). The function \(\eta_{i}\) is the link function and follows a linear model.
\(g(\mu_{i})=\eta_{i}=\mathbf{x_{i}^{\prime}}\beta\)
When inverted: \(\mu=g^{-1}(\mathbf{x_{i}^{\prime}}\beta)\)
Maximum Likelihood Estimation
For an initial rough estimate of the parameters \(\hat{\beta}\), use the estimate to generate fitted values: \(\mu_{i}=g^{-1}(\hat{\eta_{i}})\)
Let \(z\) be a working dependent variable such that \(z_{i}=\hat{\eta_{i}}+(y_{i}-\hat{\mu_{i}})\frac{d\eta_{i}}{d\mu_{i}}\),
where \(\frac{d\eta_{i}}{d\mu_{i}}\) is the derivative of the link function evaluated at the trial estimate.
Calculate the iterative weights: \(w_{i}=\frac{p_{i}}{[b^{\prime\prime}(\theta_{i})\frac{d\eta_{i}}{d\mu_{i}}^{2}]}\)
Where \(b^{\prime\prime}\) is the second derivative of \(b(\theta_{i})\) evaluated at the trial estimate.
Assume \(a_{i}(\phi)\) is of the form \(\frac{\phi}{p_{i}}\). The weight \(w_{i}\) is inversely proportional to the variance of the working dependent variable \(z_{i}\) for current parameter estimates and proportionality factor \(\phi\).
Regress \(z_{i}\) on the predictors \(x_{i}\) using the weights \(w_{i}\) to obtain new estimates of \(\beta\). \(\hat{\beta}=(\mathbf{X}^{\prime}\mathbf{W}\mathbf{X})^{-1}\mathbf{X}^{\prime}\mathbf{W}\mathbf{z}\)
Where \(\mathbf{X}\) is the model matrix, \(\mathbf{W}\) is a diagonal matrix of \(w_{i}\), and \(\mathbf{z}\) is a vector of the working response variable \(z_{i}\).
This process is repeated until the estimates \(\hat{\beta}\) change by less than the specified amount.
Cost of computation
H2O can process large data sets because it relies on parallel processes. Large data sets are divided into smaller data sets and processed simultaneously and the results are communicated between computers as needed throughout the process.
In GLM, data are split by rows but not by columns, because the predicted Y values depend on information in each of the predictor variable vectors. If O is a complexity function, N is the number of observations (or rows), and P is the number of predictors (or columns) then
\(Runtime\propto p^3+\frac{(N*p^2)}{CPUs}\)
Distribution reduces the time it takes an algorithm to process because it decreases N.
Relative to P, the larger that (N/CPUs) becomes, the more trivial p becomes to the overall computational cost. However, when p is greater than (N/CPUs), O is dominated by p.
\(Complexity = O(p^3 + N*p^2)\)
For more information about how GLM works, refer to the Generalized Linear Modeling booklet.
References
Breslow, N E. “Generalized Linear Models: Checking Assumptions and Strengthening Conclusions.” Statistica Applicata 8 (1996): 23-41.
Frome, E L. “The Analysis of Rates Using Poisson Regression Models.” Biometrics (1983): 665-674.
Snee, Ronald D. “Validation of Regression Models: Methods and Examples.” Technometrics 19.4 (1977): 415-428.
DRF
- Introduction
- Defining a DRF Model
- Interpreting a DRF Model
- Leaf Node Assignment
- FAQ
- DRF Algorithm
- References
Introduction
Distributed Random Forest (DRF) is a powerful classification tool. When given a set of data, DRF generates a forest of classification trees, rather than a single classification tree. Each of these trees is a weak learner built on a subset of rows and columns. More trees will reduce the variance. The classification from each H2O tree can be thought of as a vote; the most votes determines the classification.
The current version of DRF is fundamentally the same as in previous versions of H2O (same algorithmic steps, same histogramming techniques), with the exception of the following changes:
- Improved ability to train on categorical variables (using the
nbins_catsparameter) - Minor changes in histogramming logic for some corner cases
- By default, DRF now builds half as many trees for binomial problems, similar to GBM: one tree to estimate class 0, probability p0, class 1 probability is 1-p0.
There was some code cleanup and refactoring to support the following features:
- Per-row observation weights
- Per-row offsets
- N-fold cross-validation
DRF no longer has a special-cased histogram for classification (class DBinomHistogram has been superseded by DRealHistogram), since it was not applicable to cases with observation weights or for cross-validation.
Defining a DRF Model
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
nfolds: Specify the number of folds for cross-validation.
response_column: (Required) Select the column to use as the independent variable. The data can be numeric or categorical.
Ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
ntrees: Specify the number of trees.
max_depth: Specify the maximum tree depth.
min_rows: Specify the minimum number of observations for a leaf (
nodesizein R).nbins: (Numerical/real/int only) Specify the number of bins for the histogram to build, then split at the best point.
nbins_cats: (Categorical/enums only) Specify the maximum number of bins for the histogram to build, then split at the best point. Higher values can lead to more overfitting. The levels are ordered alphabetically; if there are more levels than bins, adjacent levels share bins. This value has a more significant impact on model fitness than nbins. Larger values may increase runtime, especially for deep trees and large clusters, so tuning may be required to find the optimal value for your configuration.
seed: Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations.
mtries: Specify the columns to randomly select at each level. If the default value of
-1is used, the number of variables is the square root of the number of columns for classification and p/3 for regression (where p is the number of predictors). The range is -1 to >=1.sample_rate: Specify the row sampling rate (x-axis). The range is 0.0 to 1.0. Higher values may improve training accuracy. Test accuracy improves when either columns or rows are sampled. For details, refer to “Stochastic Gradient Boosting” (Friedman, 1999).
col_sample_rate: Specify the column sampling rate (y-axis). The range is 0.0 to 1.0. Higher values may improve training accuracy. Test accuracy improves when either columns or rows are sampled. For details, refer to “Stochastic Gradient Boosting” (Friedman, 1999).
score_each_iteration: (Optional) Check this checkbox to score during each iteration of the model training.
score_tree_interval: Score the model after every so many trees. Disabled if set to 0.
fold_assignment: (Applicable only if a value for nfolds is specified and fold_column is not selected) Select the cross-validation fold assignment scheme. The available options are AUTO (which is Random), Random, or Modulo.
fold_column: Select the column that contains the cross-validation fold index assignment per observation.
offset_column: Select a column to use as the offset.
Note: Offsets are per-row “bias values” that are used during model training. For Gaussian distributions, they can be seen as simple corrections to the response (y) column. Instead of learning to predict the response (y-row), the model learns to predict the (row) offset of the response column. For other distributions, the offset corrections are applied in the linearized space before applying the inverse link function to get the actual response values. For more information, refer to the following link.
weights_column: Select a column to use for the observation weights, which are used for bias correction. The specified
weights_columnmust be included in the specifiedtraining_frame. Python only: To use a weights column when passing an H2OFrame toxinstead of a list of column names, the specifiedtraining_framemust contain the specifiedweights_column.Note: Weights are per-row observation weights and do not increase the size of the data frame. This is typically the number of times a row is repeated, but non-integer values are supported as well. During training, rows with higher weights matter more, due to the larger loss function pre-factor.
balance_classes: Oversample the minority classes to balance the class distribution. This option is not selected by default and can increase the data frame size. This option is only applicable for classification.
max_confusion_matrix_size: Specify the maximum size (in number of classes) for confusion matrices to be printed in the Logs.
max_hit_ratio_k: Specify the maximum number (top K) of predictions to use for hit ratio computation. Applicable to multi-class only. To disable, enter 0.
r2_stopping: Specify a threshold for the coefficient of determination (\(r^2\)) metric value. When this threshold is met or exceeded, H2O stops making trees.
stopping_rounds: Stops training when the option selected for stopping_metric doesn’t improve for the specified number of training rounds, based on a simple moving average. To disable this feature, specify
0. The metric is computed on the validation data (if provided); otherwise, training data is used. When used with overwrite_with_best_model, the final model is the best model generated for the given stopping_metric option.Note: If cross-validation is enabled:
- All cross-validation models stop training when the validation metric doesn’t improve.
- The main model runs for the mean number of epochs.
- N+1 models do not use overwrite_with_best_model
- N+1 models may be off by the number specified for stopping_rounds from the best model, but the cross-validation metric estimates the performance of the main model for the resulting number of epochs (which may be fewer than the specified number of epochs).
stopping_metric: Select the metric to use for early stopping. The available options are:
- AUTO: Logloss for classification, deviance for regression
- deviance
- logloss
- MSE
- AUC
- r2
- misclassification
stopping_tolerance: Specify the relative tolerance for the metric-based stopping to stop training if the improvement is less than this value.
max_runtime_secs: Maximum allowed runtime in seconds for model training. Use 0 to disable.
build_tree_one_node: To run on a single node, check this checkbox. This is suitable for small datasets as there is no network overhead but fewer CPUs are used.
binomial_double_trees: (Binary classification only) Build twice as many trees (one per class). Enabling this option can lead to higher accuracy, while disabling can result in faster model building. This option is disabled by default.
checkpoint: Enter a model key associated with a previously-trained model. Use this option to build a new model as a continuation of a previously-generated model.
keep_cross_validation_predictions: To keep the cross-validation predictions, check this checkbox.
class_sampling_factors: Specify the per-class (in lexicographical order) over/under-sampling ratios. By default, these ratios are automatically computed during training to obtain the class balance.
max_after_balance_size: Specify the maximum relative size of the training data after balancing class counts (balance_classes must be enabled). The value can be less than 1.0.
nbins_top_level: (For numerical/real/int columns only) Specify the minimum number of bins at the root level to use to build the histogram. This number will then be decreased by a factor of two per level.
Interpreting a DRF Model
By default, the following output displays:
- Model parameters (hidden)
- A graph of the scoring history (number of trees vs. training MSE)
- A graph of the ROC curve (TPR vs. FPR)
- A graph of the variable importances
- Output (model category, validation metrics, initf)
- Model summary (number of trees, min. depth, max. depth, mean depth, min. leaves, max. leaves, mean leaves)
- Scoring history in tabular format
- Training metrics (model name, checksum name, frame name, frame checksum name, description, model category, duration in ms, scoring time, predictions, MSE, R2, logloss, AUC, GINI)
- Training metrics for thresholds (thresholds, F1, F2, F0Points, Accuracy, Precision, Recall, Specificity, Absolute MCC, min. per-class accuracy, TNS, FNS, FPS, TPS, IDX)
- Maximum metrics (metric, threshold, value, IDX)
- Variable importances in tabular format
Leaf Node Assignment
Trees cluster observations into leaf nodes, and this information can be useful for feature engineering or model interpretability. Use h2o.predict_leaf_node_assignment\(model, frame\) to get an H2OFrame with the leaf node assignments, or click the checkbox when making predictions from Flow. Those leaf nodes represent decision rules that can be fed to other models (i.e., GLM with lambda search and strong rules) to obtain a limited set of the most important rules.
FAQ
How does the algorithm handle missing values during training?
Missing values affect tree split points. NAs always “go left”, and hence affect the split-finding math (since the corresponding response for the row still matters). If the response is missing, then the row won’t affect the split-finding math.
How does the algorithm handle missing values during testing?
During scoring, missing values “always go left” at any decision point in a tree. Due to dynamic binning in DRF, a row with a missing value typically ends up in the “leftmost bin” - with other outliers.
What happens if the response has missing values?
No errors will occur, but nothing will be learned from rows containing missing the response.
Does it matter if the data is sorted?
No.
Should data be shuffled before training?
No.
How does the algorithm handle highly imbalanced data in a response column?
Specify
balance_classes,class_sampling_factorsandmax_after_balance_sizeto control over/under-sampling.What if there are a large number of columns?
DRFs are best for datasets with fewer than a few thousand columns.
What if there are a large number of categorical factor levels?
Large numbers of categoricals are handled very efficiently - there is never any one-hot encoding.
How is variable importance calculated for DRF?
Variable importance is determined by calculating the relative influence of each variable: whether that variable was selected during splitting in the tree building process and how much the squared error (over all trees) improved as a result.
- How is column sampling implemented for DRF?
For an example model using:
- 100 columns
col_sample_rate_per_treeis 0.602mtriesis -1 or 7 (refers to the number of active predictor columns for the dataset)
For each tree, the floor is used to determine the number - for this example, (0.602*100)=60 out of the 100 - of columns that are randomly picked. For classification cases where mtries=-1, the square root - for this example, (100)=10 columns - are then randomly chosen for each split decision (out of the total 60).
For regression, the floor - in this example, (100/3)=33 columns - is used for each split by default. If mtries=7, then 7 columns are picked for each split decision (out of the 60).
mtries is configured independently of col_sample_rate_per_tree, but it can be limited by it. For example, if col_sample_rate_per_tree=0.01, then there’s only one column left for each split, regardless of how large the value for mtries is.
DRF Algorithm
References
Naïve Bayes
- Introduction
- Defining a Naïve Bayes Model
- Interpreting a Naïve Bayes Model
- FAQ
- Naïve Bayes Algorithm
- References
Introduction
Naïve Bayes (NB) is a classification algorithm that relies on strong assumptions of the independence of covariates in applying Bayes Theorem. NB models are commonly used as an alternative to decision trees for classification problems.
Defining a Naïve Bayes Model
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
response_column: (Required) Select the column to use as the independent variable. The data must be categorical and must contain at least two unique categorical levels.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
laplace: Specify the Laplace smoothing parameter. The value must be an integer >= 0.
min_sdev: Specify the minimum standard deviation to use for observations without enough data. The value must be at least 1e-10.
eps_sdev: Specify the threshold for standard deviation. The value must be positive. If this threshold is not met, the min_sdev value is used.
min_prob: Specify the minimum probability to use for observations without enough data.
eps_prob: Specify the threshold for standard deviation. If this threshold is not met, the min_sdev value is used.
compute_metrics: To compute metrics on training data, check this checkbox. The Naïve Bayes classifier assumes independence between predictor variables conditional on the response, and a Gaussian distribution of numeric predictors with mean and standard deviation computed from the training dataset. When building a Naïve Bayes classifier, every row in the training dataset that contains at least one NA will be skipped completely. If the test dataset has missing values, then those predictors are omitted in the probability calculation during prediction.
score_each_iteration: (Optional) Check this checkbox to score during each iteration of the model training.
max_confusion_matrix_size: Specify the maximum size (in number of classes) for confusion matrices to be printed in the Logs.
max_hit_ratio_k: Specify the maximum number (top K) of predictions to use for hit ratio computation. Applicable to multi-class only. To disable, enter 0.
max_runtime_secs: Maximum allowed runtime in seconds for model training. Use 0 to disable.
Interpreting a Naïve Bayes Model
The output from Naïve Bayes is a list of tables containing the a-priori and conditional probabilities of each class of the response. The a-priori probability is the estimated probability of a particular class before observing any of the predictors. Each conditional probability table corresponds to a predictor column. The row headers are the classes of the response and the column headers are the classes of the predictor. Thus, in the table below, the probability of survival (y) given a person is male (x) is 0.91543624.
Sex
Survived Male Female
No 0.91543624 0.08456376
Yes 0.51617440 0.48382560
When the predictor is numeric, Naïve Bayes assumes it is sampled from a Gaussian distribution given the class of the response. The first column contains the mean and the second column contains the standard deviation of the distribution.
By default, the following output displays:
- Output (model category, model summary, scoring history, training metrics, validation metrics)
- Y-Levels (levels of the response column)
- P-conditionals
FAQ
How does the algorithm handle missing values during training?
All rows with one or more missing values (either in the predictors or the response) will be skipped during model building.
How does the algorithm handle missing values during testing?
If a predictor is missing, it will be skipped when taking the product of conditional probabilities in calculating the joint probability conditional on the response.
What happens if the response domain is different in the training and test datasets?
The response column in the test dataset is not used during scoring, so any response categories absent in the training data will not be predicted.
What happens during prediction if the new sample has categorical levels not seen in training?
The conditional probability of that predictor level will be set according to the Laplace smoothing factor. If Laplace smoothing is disabled (set to zero), the joint probability will be zero. See pgs. 13-14 of Andrew Ng’s “Generative learning algorithms” in the References section for mathematical details.
Does it matter if the data is sorted?
No.
Should data be shuffled before training?
This does not affect model building.
How does the algorithm handle highly imbalanced data in a response column?
Unbalanced data will not affect the model. However, if one response category has very few observations compared to the total, the conditional probability may be very low. A cutoff (
eps_prob) and minimum value (min_prob) are available for the user to set a floor on the calculated probability.
What if there are a large number of columns?
More memory will be allocated on each node to store the joint frequency counts and sums.
What if there are a large number of categorical factor levels?
More memory will be allocated on each node to store the joint frequency count of each categorical predictor level with the response’s level.
When running PCA, is it better to create a cluster that uses many smaller nodes or fewer larger nodes?
For Naïve Bayes, we recommend using many smaller nodes because the distributed task doesn’t require intensive computation.
Naïve Bayes Algorithm
The algorithm is presented for the simplified binomial case without loss of generality.
Under the Naive Bayes assumption of independence, given a training set for a set of discrete valued features X \({(X^{(i)},\ y^{(i)};\ i=1,...m)}\)
The joint likelihood of the data can be expressed as:
\(\mathcal{L} \: (\phi(y),\: \phi_{i|y=1},\:\phi_{i|y=0})=\Pi_{i=1}^{m} p(X^{(i)},\: y^{(i)})\)
The model can be parameterized by:
\(\phi_{i|y=0}=\ p(x_{i}=1|\ y=0);\: \phi_{i|y=1}=\ p(x_{i}=1|y=1);\: \phi(y)\)
Where \(\phi_{i|y=0}=\ p(x_{i}=1|\ y=0)\) can be thought of as the fraction of the observed instances where feature \(x_{i}\) is observed, and the outcome is \(y=0, \phi_{i|y=1}=p(x_{i}=1|\ y=1)\) is the fraction of the observed instances where feature \(x_{i}\) is observed, and the outcome is \(y=1\), and so on.
The objective of the algorithm is to maximize with respect to \(\phi_{i|y=0}, \ \phi_{i|y=1},\ and \ \phi(y)\)
Where the maximum likelihood estimates are:
\(\phi_{j|y=1}= \frac{\Sigma_{i}^m 1(x_{j}^{(i)}=1 \ \bigcap y^{i} = 1)}{\Sigma_{i=1}^{m}(y^{(i)}=1}\)
\(\phi_{j|y=0}= \frac{\Sigma_{i}^m 1(x_{j}^{(i)}=1 \ \bigcap y^{i} = 0)}{\Sigma_{i=1}^{m}(y^{(i)}=0}\)
\(\phi(y)= \frac{(y^{i} = 1)}{m}\)
Once all parameters \(\phi_{j|y}\) are fitted, the model can be used to predict new examples with features \(X_{(i^*)}\).
This is carried out by calculating:
\(p(y=1|x)=\frac{\Pi p(x_i|y=1) p(y=1)}{\Pi p(x_i|y=1)p(y=1) \: +\: \Pi p(x_i|y=0)p(y=0)}\)
\(p(y=0|x)=\frac{\Pi p(x_i|y=0) p(y=0)}{\Pi p(x_i|y=1)p(y=1) \: +\: \Pi p(x_i|y=0)p(y=0)}\)
and predicting the class with the highest probability.
It is possible that prediction sets contain features not originally seen in the training set. If this occurs, the maximum likelihood estimates for these features predict a probability of 0 for all cases of y.
Laplace smoothing allows a model to predict on out of training data features by adjusting the maximum likelihood estimates to be:
\(\phi_{j|y=1}= \frac{\Sigma_{i}^m 1(x_{j}^{(i)}=1 \ \bigcap y^{i} = 1) \: + \: 1}{\Sigma_{i=1}^{m}(y^{(i)}=1 \: + \: 2}\)
\(\phi_{j|y=0}= \frac{\Sigma_{i}^m 1(x_{j}^{(i)}=1 \ \bigcap y^{i} = 0) \: + \: 1}{\Sigma_{i=1}^{m}(y^{(i)}=0 \: + \: 2}\)
Note that in the general case where y takes on k values, there are k+1 modified parameter estimates, and they are added in when the denominator is k (rather than two, as shown in the two-level classifier shown here.)
Laplace smoothing should be used with care; it is generally intended to allow for predictions in rare events. As prediction data becomes increasingly distinct from training data, train new models when possible to account for a broader set of possible X values.
References
Ng, Andrew. “Generative Learning algorithms.” (2008).
PCA
Introduction
Principal Components Analysis (PCA) is closely related to Principal Components Regression. The algorithm is carried out on a set of possibly collinear features and performs a transformation to produce a new set of uncorrelated features.
PCA is commonly used to model without regularization or perform dimensionality reduction. It can also be useful to carry out as a preprocessing step before distance-based algorithms such as K-Means since PCA guarantees that all dimensions of a manifold are orthogonal.
Defining a PCA Model
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
transform: Select the transformation method for the training data: None, Standardize, Normalize, Demean, or Descale. The default is None.
pca_method: Select the algorithm to use for computing the principal components:
- GramSVD: Uses a distributed computation of the Gram matrix, followed by a local SVD using the JAMA package
- Power: Computes the SVD using the power iteration method (experimental)
- Randomized: Uses randomized subspace iteration method
- GLRM: Fits a generalized low-rank model with L2 loss function and no regularization and solves for the SVD using local matrix algebra (experimental)
k*: Specify the rank of matrix approximation. The default is 1.
max_iterations: Specify the number of training iterations. The value must be between 1 and 1e6 and the default is 1000.
seed: Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations.
use_all_factor_levels: Check this checkbox to use all factor levels in the possible set of predictors; if you enable this option, sufficient regularization is required. By default, the first factor level is skipped. For PCA models, this option ignores the first factor level of each categorical column when expanding into indicator columns.
compute_metrics: Enable metrics computations on the training data.
score_each_iteration: (Optional) Check this checkbox to score during each iteration of the model training.
max_runtime_secs: Maximum allowed runtime in seconds for model training. Use 0 to disable.
Interpreting a PCA Model
PCA output returns a table displaying the number of components specified by the value for k.
Scree and cumulative variance plots for the components are returned as well. Users can access them by clicking on the black button labeled “Scree and Variance Plots” at the top left of the results page. A scree plot shows the variance of each component, while the cumulative variance plot shows the total variance accounted for by the set of components.
The output for PCA includes the following:
- Model parameters (hidden)
- Output (model category, model summary, scoring history, training metrics, validation metrics, iterations)
- Archetypes
- Standard deviation
- Rotation
- Importance of components (standard deviation, proportion of variance, cumulative proportion)
FAQ
- How does the algorithm handle missing values during scoring?
For the GramSVD and Power methods, all rows containing missing values are ignored during training. For the GLRM method, missing values are excluded from the sum over the loss function in the objective. For more information, refer to section 4 Generalized Loss Functions, equation (13), in “Generalized Low Rank Models” by Boyd et al.
How does the algorithm handle missing values during testing?
During scoring, the test data is right-multiplied by the eigenvector matrix produced by PCA. Missing categorical values are skipped in the row product-sum. Missing numeric values propagate an entire row of NAs in the resulting projection matrix.
What happens during prediction if the new sample has categorical levels not seen in training?
Categorical levels in the test data not present in the training data are skipped in the row product-sum.
Does it matter if the data is sorted?
No, sorting data does not affect the model.
Should data be shuffled before training?
No, shuffling data does not affect the model.
What if there are a large number of columns?
Calculating the SVD will be slower, since computations on the Gram matrix are handled locally.
What if there are a large number of categorical factor levels?
Each factor level (with the exception of the first, depending on whether use_all_factor_levels is enabled) is assigned an indicator column. The indicator column is 1 if the observation corresponds to a particular factor; otherwise, it is 0. As a result, many factor levels result in a large Gram matrix and slower computation of the SVD.
How are categorical columns handled during model building?
If the GramSVD or Power methods are used, the categorical columns are expanded into 0/1 indicator columns for each factor level. The algorithm is then performed on this expanded training frame. For GLRM, the multidimensional loss function for categorical columns is discussed in Section 6.1 of “Generalized Low Rank Models” by Boyd et al.
When running PCA, is it better to create a cluster that uses many smaller nodes or fewer larger nodes?
For PCA, this is dependent on the selected pca_method parameter:
- For GramSVD, use fewer larger nodes for better performance. Forming the Gram matrix requires few intensive calculations and the main bottleneck is the JAMA library’s SVD function, which is not parallelized and runs on a single machine. We do not recommend selecting GramSVD for datasets with many columns and/or categorical levels in one or more columns.
- For Randomized, use many smaller nodes for better performance, since H2O calls a few different distributed tasks in a loop, where each task does fairly simple matrix algebra computations.
- For GLRM, the number of nodes depends on whether the dataset contains many categorical columns with many levels. If this is the case, we recommend using fewer larger nodes, since computing the loss function for categoricals is an intensive task. If the majority of the data is numeric and the categorical columns have only a small number of levels (~10-20), we recommend using many small nodes in the cluster.
For Power, we recommend using fewer larger nodes because the intensive calculations are single-threaded. However, this method is only recommended for obtaining principal component values (such as
k << ncol(train))because the other methods are far more efficient.I ran PCA on my dataset - how do I input the new parameters into a model?
After the PCA model has been built using h2o.prcomp, use h2o.predict on the original data frame and the PCA model to produce the dimensionality-reduced representation. Use cbind to add the predictor column from the original data frame to the data frame produced by the output of h2o.predict. At this point, you can build supervised learning models on the new data frame.
PCA Algorithm
Let \(X\) be an \(M\times N\) matrix where
Each row corresponds to the set of all measurements on a particular attribute, and
Each column corresponds to a set of measurements from a given observation or trial
The covariance matrix \(C_{x}\) is
\(C_{x}=\frac{1}{n}XX^{T}\)
where \(n\) is the number of observations.
\(C_{x}\) is a square, symmetric \(m\times m\) matrix, the diagonal entries of which are the variances of attributes, and the off-diagonal entries are covariances between attributes.
PCA convergence is based on the method described by Gockenbach: “The rate of convergence of the power method depends on the ratio \(lambda_2|/|\lambda_1\). If this is small…then the power method converges rapidly. If the ratio is close to 1, then convergence is quite slow. The power method will fail if \(lambda_2| = |\lambda_1\).” (567).
The objective of PCA is to maximize variance while minimizing covariance.
To accomplish this, for a new matrix \(C_{y}\) with off diagonal entries of 0, and each successive dimension of Y ranked according to variance, PCA finds an orthonormal matrix \(P\) such that \(Y=PX\) constrained by the requirement that \(C_{y}=\frac{1}{n}YY^{T}\) be a diagonal matrix.
The rows of \(P\) are the principal components of X.
\(C_{y}=\frac{1}{n}YY^{T}\) \(=\frac{1}{n}(PX)(PX)^{T}\) \(C_{y}=PC_{x}P^{T}.\)
Because any symmetric matrix is diagonalized by an orthogonal matrix of its eigenvectors, solve matrix \(P\) to be a matrix where each row is an eigenvector of \(\frac{1}{n}XX^{T}=C_{x}\)
Then the principal components of \(X\) are the eigenvectors of \(C_{x}\), and the \(i^{th}\) diagonal value of \(C_{y}\) is the variance of \(X\) along \(p_{i}\).
Eigenvectors of \(C_{x}\) are found by first finding the eigenvalues \(\lambda\) of \(C_{x}\).
For each eigenvalue \(\lambda\) \((C-{x}-\lambda I)x =0\) where \(x\) is the eigenvector associated with \(\lambda\).
Solve for \(x\) by Gaussian elimination.
Recovering SVD from GLRM
GLRM gives \(x\) and \(y\), where \(x \in \rm \Bbb I \!\Bbb R ^{n * k}\) and \( y \in \rm \Bbb I \!\Bbb R ^{k*m} \)
- \(n\)= number of rows (A)
- \(m\)= number of columns (A)
- \(k\)= user-specified rank - \(A\)= training matrix
It is assumed that the \(x\) and \(y\) columns are independent.
First, perform QR decomposition of \(x\) and \(y^T\):
\(x = QR\)
\(y^T = ZS\), where \(Q^TQ = I = Z^TZ\)
Call JAMA QR Decomposition directly on \(y^T\) to get \( Z \in \rm \Bbb I \! \Bbb R\), \( S \in \Bbb I \! \Bbb R \)
\( R \) from QR decomposition of \( x \) is the upper triangular factor of Cholesky of \(X^TX\) Gram
\( X^TX = LL^T, X = QR \)
\( X^TX= (R^TQ^T) QR = R^TR \), since \(Q^TQ=I \) => \(R=L^T\) (transpose lower triangular)
Note: In code, \(X^TX \over n\) = \( LL^T \)
\( X^TX = (L \sqrt{n})(L \sqrt{n})^T =R^TR \)
\( R = L^T \sqrt{n} \in \rm \Bbb I \! \Bbb R^{k * k} \) reduced QR decomposition.
For more information, refer to the Rectangular matrix section of “QR Decomposition” on Wikipedia.
\( XY = QR(ZS)^T = Q(RS^T)Z^T \)
Note: \( (RS^T) \in \rm \Bbb I \!\Bbb R \)
Find SVD (locally) of \( RS^T \)
\( RS^T = U \sum V^T, U^TU = I = V^TV \) orthogonal
\( XY = Q(RS^T)Z^T = (QU \sum (V^T Z^T) SVD \)
\( (QU)^T(QU) = U^T Q^TQU U^TU = I\)
\( (ZV)^T(ZV) = V^TZ^TZV = V^TV =I \)
Right singular vectors: \( ZV \in \rm \Bbb I \!\Bbb R^{m * k} \)
Singular values: \( \sum \in \rm \Bbb I \!\Bbb R^{k * k} \) diagonal
Left singular vectors: \( (QU) \in \rm \Bbb I \!\Bbb R^{n * k}\)
References
Gockenbach, Mark S. “Finite-Dimensional Linear Algebra (Discrete Mathematics and Its Applications).” (2010): 566-567.
GBM
- Introduction
- Defining a GBM Model
- Interpreting a GBM Model
- Leaf Node Assignment
- FAQ
- GBM Algorithm
- Binning In GBM
- References
Introduction
Gradient Boosted Regression and Gradient Boosted Classification are forward learning ensemble methods. The guiding heuristic is that good predictive results can be obtained through increasingly refined approximations. H2O’s GBM sequentially builds regression trees on all the features of the dataset in a fully distributed way - each tree is built in parallel.
The current version of GBM is fundamentally the same as in previous versions of H2O (same algorithmic steps, same histogramming techniques), with the exception of the following changes:
- Improved ability to train on categorical variables (using the
nbins_catsparameter) - Minor changes in histogramming logic for some corner cases
There was some code cleanup and refactoring to support the following features:
- Per-row observation weights
- Per-row offsets
- N-fold cross-validation
- Support for more distribution functions (such as Gamma, Poisson, and Tweedie)
Defining a GBM Model
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
nfolds: Specify the number of folds for cross-validation.
response_column: (Required) Select the column to use as the independent variable. The data can be numeric or categorical.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
ntrees: Specify the number of trees.
max_depth: Specify the maximum tree depth.
min_rows: Specify the minimum number of observations for a leaf (
nodesizein R).nbins: (Numerical/real/int only) Specify the number of bins for the histogram to build, then split at the best point.
nbins_cats: (Categorical/enums only) Specify the maximum number of bins for the histogram to build, then split at the best point. Higher values can lead to more overfitting. The levels are ordered alphabetically; if there are more levels than bins, adjacent levels share bins. This value has a more significant impact on model fitness than nbins. Larger values may increase runtime, especially for deep trees and large clusters, so tuning may be required to find the optimal value for your configuration.
seed: Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations.
learn_rate: Specify the learning rate. The range is 0.0 to 1.0.
distribution: Select the loss function. The options are auto, bernoulli, multinomial, gaussian, poisson, gamma, or tweedie.
- If the distribution is multinomial, the response column must be categorical.
- If the distribution is poisson, the response column must be numeric.
- If the distribution is gamma, the response column must be numeric.
- If the distribution is tweedie, the response column must be numeric.
- If the distribution is gaussian, the response column must be numeric.
- If the distribution is laplace, the data must be numeric and continuous (Int).
- If the distribution is quantile, the data must be numeric and continuous (Int).
sample_rate: Specify the row sampling rate (x-axis). The range is 0.0 to 1.0. Higher values may improve training accuracy. Test accuracy improves when either columns or rows are sampled. For details, refer to “Stochastic Gradient Boosting” (Friedman, 1999).
col_sample_rate: Specify the column sampling rate (y-axis). The range is 0.0 to 1.0. Higher values may improve training accuracy. Test accuracy improves when either columns or rows are sampled. For details, refer to “Stochastic Gradient Boosting” (Friedman, 1999).
score_each_iteration: (Optional) Check this checkbox to score during each iteration of the model training.
score_tree_interval: Score the model after every so many trees. Disabled if set to 0.
fold_assignment: (Applicable only if a value for nfolds is specified and fold_column is not selected) Select the cross-validation fold assignment scheme. The available options are AUTO (which is Random), Random, or Modulo.
fold_column: Select the column that contains the cross-validation fold index assignment per observation.
offset_column: (Not applicable if the distribution is multinomial) Select a column to use as the offset.
Note: Offsets are per-row “bias values” that are used during model training. For Gaussian distributions, they can be seen as simple corrections to the response (y) column. Instead of learning to predict the response (y-row), the model learns to predict the (row) offset of the response column. For other distributions, the offset corrections are applied in the linearized space before applying the inverse link function to get the actual response values. For more information, refer to the following link. If the distribution is Bernoulli, the value must be less than one.
weights_column: Select a column to use for the observation weights, which are used for bias correction. The specified
weights_columnmust be included in the specifiedtraining_frame. Python only: To use a weights column when passing an H2OFrame toxinstead of a list of column names, the specifiedtraining_framemust contain the specifiedweights_column.Note: Weights are per-row observation weights and do not increase the size of the data frame. This is typically the number of times a row is repeated, but non-integer values are supported as well. During training, rows with higher weights matter more, due to the larger loss function pre-factor.
balance_classes: Oversample the minority classes to balance the class distribution. This option is not selected by default and can increase the data frame size. This option is only applicable for classification. Majority classes can be undersampled to satisfy the Max_after_balance_size parameter.
max_confusion_matrix_size: Specify the maximum size (in number of classes) for confusion matrices to be printed in the Logs.
max_hit_ratio_k: Specify the maximum number (top K) of predictions to use for hit ratio computation. Applicable to multi-class only. To disable, enter 0.
r2_stopping: Specify a threshold for the coefficient of determination (\(r^2\)) metric value. When this threshold is met or exceeded, H2O stops making trees.
stopping_rounds: Stops training when the option selected for stopping_metric doesn’t improve for the specified number of training rounds, based on a simple moving average. To disable this feature, specify
0. The metric is computed on the validation data (if provided); otherwise, training data is used. When used with overwrite_with_best_model, the final model is the best model generated for the given stopping_metric option.Note: If cross-validation is enabled:
- All cross-validation models stop training when the validation metric doesn’t improve.
- The main model runs for the mean number of epochs.
- N+1 models do not use overwrite_with_best_model
- N+1 models may be off by the number specified for stopping_rounds from the best model, but the cross-validation metric estimates the performance of the main model for the resulting number of epochs (which may be fewer than the specified number of epochs).
stopping_metric: Select the metric to use for early stopping. The available options are:
- AUTO: Logloss for classification, deviance for regression
- deviance
- logloss
- MSE
- AUC
- r2
- misclassification
stopping_tolerance: Specify the relative tolerance for the metric-based stopping to stop training if the improvement is less than this value.
max_runtime_secs: Maximum allowed runtime in seconds for model training. Use 0 to disable.
build_tree_one_node: To run on a single node, check this checkbox. This is suitable for small datasets as there is no network overhead but fewer CPUs are used.
quantile_alpha: (Only applicable if Quantile is selected for distribution) Specify the quantile to be used for Quantile Regression.
tweedie_power: (Only applicable if Tweedie is selected for distribution) Specify the Tweedie power. The range is from 1 to 2. For a normal distribution, enter
0. For Poisson distribution, enter1. For a gamma distribution, enter2. For a compound Poisson-gamma distribution, enter a value greater than 1 but less than 2. For more information, refer to Tweedie distribution.checkpoint: Enter a model key associated with a previously-trained model. Use this option to build a new model as a continuation of a previously-generated model.
keep_cross_validation_predictions: To keep the cross-validation predictions, check this checkbox.
class_sampling_factors: Specify the per-class (in lexicographical order) over/under-sampling ratios. By default, these ratios are automatically computed during training to obtain the class balance. There is no default value.
max_after_balance_size: Specify the maximum relative size of the training data after balancing class counts (balance_classes must be enabled). The value can be less than 1.0.
nbins_top_level: (For numerical/real/int columns only) Specify the minimum number of bins at the root level to use to build the histogram. This number will then be decreased by a factor of two per level.
Interpreting a GBM Model
The output for GBM includes the following:
- Model parameters (hidden)
- A graph of the scoring history (training MSE vs number of trees)
- A graph of the variable importances
- Output (model category, validation metrics, initf)
- Model summary (number of trees, min. depth, max. depth, mean depth, min. leaves, max. leaves, mean leaves)
- Scoring history in tabular format
- Training metrics (model name, model checksum name, frame name, description, model category, duration in ms, scoring time, predictions, MSE, R2)
- Variable importances in tabular format
Leaf Node Assignment
Trees cluster observations into leaf nodes, and this information can be useful for feature engineering or model interpretability. Use h2o.predict_leaf_node_assignment\(model, frame\) to get an H2OFrame with the leaf node assignments, or click the checkbox when making predictions from Flow. Those leaf nodes represent decision rules that can be fed to other models (i.e., GLM with lambda search and strong rules) to obtain a limited set of the most important rules.
FAQ
How does the algorithm handle missing values during training?
Missing values affect tree split points. NAs always “go left”, and hence affect the split-finding math (since the corresponding response for the row still matters). If the response is missing, then the row won’t affect the split-finding math.
How does the algorithm handle missing values during testing?
During scoring, missing values “always go left” at any decision point in a tree. Due to dynamic binning in GBM, a row with a missing value typically ends up in the “leftmost bin” - with other outliers.
What happens if the response has missing values?
No errors will occur, but nothing will be learned from rows containing missing the response.
Does it matter if the data is sorted?
No.
Should data be shuffled before training?
No.
How does the algorithm handle highly imbalanced data in a response column?
You can specify
balance_classes,class_sampling_factorsandmax_after_balance_sizeto control over/under-sampling.What if there are a large number of columns?
DRF models are best for datasets with fewer than a few thousand columns.
What if there are a large number of categorical factor levels?
Large numbers of categoricals are handled very efficiently - there is never any one-hot encoding.
Given the same training set and the same GBM parameters, will GBM produce a different model with two different validation data sets, or the same model?
The same model will be generated.
How deterministic is GBM?
The
nfoldsandbalance_classesparameters use the seed directly. Otherwise, GBM is deterministic up to floating point rounding errors (out-of-order atomic addition of multiple threads during histogram building). Any observed variations in the AUC curve should be the same up to at least three to four significant digits.When fitting a random number between 0 and 1 as a single feature, the training ROC curve is consistent with
randomfor low tree numbers and overfits as the number of trees is increased, as expected. However, when a random number is included as part of a set of hundreds of features, as the number of trees increases, the random number increases in feature importance. Why is this?
This is a known behavior of GBM that is similar to its behavior in R. If, for example, it takes 50 trees to learn all there is to learn from a frame without the random features, when you add a random predictor and train 1000 trees, the first 50 trees will be approximately the same. The final 950 trees are used to make sense of the random number, which will take a long time since there’s no structure. The variable importance will reflect the fact that all the splits from the first 950 trees are devoted to the random feature.
- How is column sampling implemented for GBM?
For an example model using:
- 100 columns
col_sample_rate_per_tree=0.754col_sample_rate=0.8(refers to available columns after per-tree sampling)
For each tree, the floor is used to determine the number - in this example, (0.754100)=75 out of the 100 - of columns that are randomly picked, and then the floor is used to determine the number - in this case,(0.7540.8*100)=60 - of columns that are then randomly chosen for each split decision (out of the 75).
GBM Algorithm
H2O’s Gradient Boosting Algorithms follow the algorithm specified by Hastie et al (2001):
Initialize \(f_{k0} = 0,\: k=1,2,…,K\)
For \(m=1\) to \(M:\)
(a) Set \(p_{k}(x)=\frac{e^{f_{k}(x)}}{\sum_{l=1}^{K}e^{f_{l}(x)}},\:k=1,2,…,K\)
(b) For \(k=1\) to \(K\):
i. Compute \(r_{ikm}=y_{ik}-p_{k}(x_{i}),\:i=1,2,…,N.\) ii. Fit a regression tree to the targets \(r_{ikm},\:i=1,2,…,N\), giving terminal regions \(R_{jim},\:j=1,2,…,J_{m}.\) \(iii. Compute\) \(\gamma_{jkm}=\frac{K-1}{K}\:\frac{\sum_{x_{i}\in R_{jkm}}(r_{ikm})}{\sum_{x_{i}\in R_{jkm}}|r_{ikm}|(1-|r_{ikm})},\:j=1,2,…,J_{m}.\) \(\:iv.\:Update\:f_{km}(x)=f_{k,m-1}(x)+\sum_{j=1}^{J_{m}}\gamma_{jkm}I(x\in\:R_{jkm}).\)
Output \(\:\hat{f_{k}}(x)=f_{kM}(x),\:k=1,2,…,K.\)
Be aware that the column type affects how the histogram is created and the column type depends on whether rows are excluded or assigned a weight of 0. For example:
val weight 1 1 0.5 0 5 1 3.5 0
The above vec has a real-valued type if passed as a whole, but if the zero-weighted rows are sliced away first, the integer weight is used. The resulting histogram is either kept at full nbins resolution or potentially shrunk to the discrete integer range, which affects the split points.
For more information about the GBM algorithm, refer to the Gradient Boosted Machines booklet.
Binning In GBM
Is the binning range-based or percentile-based?
It’s range based, and re-binned at each tree split. NAs always “go to the left” (smallest) bin. There’s a minimum observations required value (default 10). There has to be at least 1 FP ULP improvement in error to split (all-constant predictors won’t split). nbins is at least 1024 at the top-level, and divides by 2 down each level until you hit the nbins parameter (default: 20). Categoricals use a separate, more aggressive, binning range.
Re-binning means, eg, suppose your column C1 data is: {1,1,2,4,8,16,100,1000}. Then a 20-way binning will use the range from 1 to 1000, bin by units of 50. The first binning will be a lumpy: {1,1,2,4,8,16},{100},{47_empty_bins},{1000}. Suppose the split peels out the {1000} bin from the rest.
Next layer in the tree for the left-split has value from 1 to 100 (not 1000!) and so re-bins in units of 5: {1,1,2,4},{8},{},{16},{lots of empty bins}{100} (the RH split has the single value 1000).
And so on: important dense ranges with split essentially logrithmeticaly at each layer.
What should I do if my variables are long skewed in the tail and might have large outliers?
You can try adding a new predictor column which is either pre-binned (e.g. as a categorical - “small”, “median”, and “giant” values), or a log-transform - plus keep the old column.
References
Dietterich, Thomas G, and Eun Bae Kong. “Machine Learning Bias, Statistical Bias, and Statistical Variance of Decision Tree Algorithms.” ML-95 255 (1995).
Elith, Jane, John R Leathwick, and Trevor Hastie. “A Working Guide to Boosted Regression Trees.” Journal of Animal Ecology 77.4 (2008): 802-813
Friedman, Jerome H. “Greedy Function Approximation: A Gradient Boosting Machine.” Annals of Statistics (2001): 1189-1232.
Friedman, Jerome, Trevor Hastie, Saharon Rosset, Robert Tibshirani, and Ji Zhu. “Discussion of Boosting Papers.” Ann. Statist 32 (2004): 102-107
Deep Learning
- Introduction
- Defining a Deep Learning Model
- Interpreting a Deep Learning Model
- FAQ
- Deep Learning Algorithm
- References
Introduction
H2O’s Deep Learning is based on a multi-layer feed-forward artificial neural network that is trained with stochastic gradient descent using back-propagation. The network can contain a large number of hidden layers consisting of neurons with tanh, rectifier and maxout activation functions. Advanced features such as adaptive learning rate, rate annealing, momentum training, dropout, L1 or L2 regularization, checkpointing and grid search enable high predictive accuracy. Each compute node trains a copy of the global model parameters on its local data with multi-threading (asynchronously), and contributes periodically to the global model via model averaging across the network.
Defining a Deep Learning Model
H2O Deep Learning models have many input parameters, many of which are only accessible via the expert mode. For most cases, use the default values. Please read the following instructions before building extensive Deep Learning models. The application of grid search and successive continuation of winning models via checkpoint restart is highly recommended, as model performance can vary greatly.
model_id: (Optional) Enter a custom name for the model to use as a reference. By default, H2O automatically generates a destination key.
training_frame: (Required) Select the dataset used to build the model. NOTE: If you click the Build a model button from the
Parsecell, the training frame is entered automatically.validation_frame: (Optional) Select the dataset used to evaluate the accuracy of the model.
nfolds: Specify the number of folds for cross-validation.
Note: Cross-validation is not supported when autoencoder is enabled.
response_column: Select the column to use as the independent variable. The data can be numeric or categorical.
ignored_columns: (Optional) Click the checkbox next to a column name to add it to the list of columns excluded from the model. To add all columns, click the All button. To remove a column from the list of ignored columns, click the X next to the column name. To remove all columns from the list of ignored columns, click the None button. To search for a specific column, type the column name in the Search field above the column list. To only show columns with a specific percentage of missing values, specify the percentage in the Only show columns with more than 0% missing values field. To change the selections for the hidden columns, use the Select Visible or Deselect Visible buttons.
ignore_const_cols: Check this checkbox to ignore constant training columns, since no information can be gained from them. This option is selected by default.
activation: Select the activation function (Tahn, Tahn with dropout, Rectifier, Rectifier with dropout, Maxout, Maxout with dropout).
- Maxout is not supported when autoencoder is enabled.
hidden: Specify the hidden layer sizes (e.g., 100,100). The value must be positive.
epochs: Specify the number of times to iterate (stream) the dataset. The value can be a fraction.
variable_importances: Check this checkbox to compute variable importance. This option is not selected by default.
fold_assignment: (Applicable only if a value for nfolds is specified and fold_column is not selected) Select the cross-validation fold assignment scheme. The available options are AUTO (which is Random), Random, or Modulo.
fold_column: Select the column that contains the cross-validation fold index assignment per observation.
weights_column: Select a column to use for the observation weights, which are used for bias correction. The specified
weights_columnmust be included in the specifiedtraining_frame. Python only: To use a weights column when passing an H2OFrame toxinstead of a list of column names, the specifiedtraining_framemust contain the specifiedweights_column.Note: Weights are per-row observation weights. This is typically the number of times a row is repeated, but non-integer values are supported as well. During training, rows with higher weights matter more, due to the larger loss function pre-factor.
offset_column: (Applicable for regression only) Select a column to use as the offset.
Note: Offsets are per-row “bias values” that are used during model training. For Gaussian distributions, they can be seen as simple corrections to the response (y) column. Instead of learning to predict the response (y-row), the model learns to predict the (row) offset of the response column. For other distributions, the offset corrections are applied in the linearized space before applying the inverse link function to get the actual response values. For more information, refer to the following link.
balance_classes: (Applicable for classification only) Oversample the minority classes to balance the class distribution. This option is not selected by default and can increase the data frame size. This option is only applicable for classification. Majority classes can be undersampled to satisfy the Max_after_balance_size parameter.
standardize: If enabled, automatically standardize the data (mean 0, variance 0). If disabled, the user must provide properly scaled input data.
max_confusion_matrix_size: Specify the maximum size (in number of classes) for confusion matrices to be printed in the Logs.
max_hit_ratio_k: Specify the maximum number (top K) of predictions to use for hit ratio computation. Applicable to multi-class only. To disable, enter 0.
checkpoint: Enter a model key associated with a previously-trained Deep Learning model. Use this option to build a new model as a continuation of a previously-generated model.
Note: Cross-validation is not supported during checkpoint restarts.
use_all_factor_levels: Check this checkbox to use all factor levels in the possible set of predictors; if you enable this option, sufficient regularization is required. By default, the first factor level is skipped. For Deep Learning models, this option is useful for determining variable importances and is automatically enabled if the autoencoder is selected.
train_samples_per_iteration: Specify the number of global training samples per MapReduce iteration. To specify one epoch, enter 0. To specify all available data (e.g., replicated training data), enter -1. To use the automatic values, enter -2.
adaptive_rate: Check this checkbox to enable the adaptive learning rate (ADADELTA). This option is selected by default.
input_dropout_ratio: Specify the input layer dropout ratio to improve generalization. Suggested values are 0.1 or 0.2.
hidden_dropout_ratios: (Applicable only if the activation type is TanhWithDropout, RectifierWithDropout, or MaxoutWithDropout) Specify the hidden layer dropout ratio to improve generalization. Specify one value per hidden layer. The range is >= 0 to <1 and the default is 0.5.
l1: Specify the L1 regularization to add stability and improve generalization; sets the value of many weights to 0.
l2: Specify the L2 regularization to add stability and improve generalization; sets the value of many weights to smaller values.
loss: Select the loss function. The options are Automatic, CrossEntropy, Quadratic, Huber, or Absolute and the default value is Automatic.
- Use Absolute, Quadratic, or Huber for regression
- Use Absolute, Quadratic, Huber, or CrossEntropy for classification
distribution: Select the distribution type from the drop-down list. The options are auto, bernoulli, multinomial, gaussian, poisson, gamma, laplace, quantile or tweedie.
quantile_alpha: (Only applicable if Quantile is selected for distribution) Specify the quantile to be used for Quantile Regression.
tweedie_power: (Only applicable if Tweedie is selected for distribution) Specify the Tweedie power. The range is from 1 to 2. For a normal distribution, enter
0. For Poisson distribution, enter1. For a gamma distribution, enter2. For a compound Poisson-gamma distribution, enter a value greater than 1 but less than 2. For more information, refer to Tweedie distribution.score_interval: Specify the shortest time interval (in seconds) to wait between model scoring.
score_training_samples: Specify the number of training set samples for scoring. The value must be >= 0. To use all training samples, enter 0.
score_validation_samples: (Applicable only if validation_frame is specified) Specify the number of validation set samples for scoring. The value must be >= 0. To use all validation samples, enter 0.
score_duty_cycle: Specify the maximum duty cycle fraction for scoring. A lower value results in more training and a higher value results in more scoring.
stopping_rounds: Stops training when the option selected for stopping_metric doesn’t improve for the specified number of training rounds, based on a simple moving average. To disable this feature, specify
0. The metric is computed on the validation data (if provided); otherwise, training data is used. When used with overwrite_with_best_model, the final model is the best model generated for the given stopping_metric option.Note: If cross-validation is enabled:
- All cross-validation models stop training when the validation metric doesn’t improve.
- The main model runs for the mean number of epochs.
- N+1 models do not use overwrite_with_best_model
- N+1 models may be off by the number specified for stopping_rounds from the best model, but the cross-validation metric estimates the performance of the main model for the resulting number of epochs (which may be fewer than the specified number of epochs).
stopping_metric: Select the metric to use for early stopping. The available options are:
- AUTO: Logloss for classification, deviance for regression
- deviance
- logloss
- MSE
- AUC
- r2
- misclassification
stopping_tolerance: Specify the relative tolerance for the metric-based stopping to stop training if the improvement is less than this value.
max_runtime_secs: Maximum allowed runtime in seconds for model training. Use 0 to disable.
autoencoder: Check this checkbox to enable the Deep Learning autoencoder. This option is not selected by default.
Note: Cross-validation is not supported when autoencoder is enabled.
keep_cross_validation_predictions: To keep the cross-validation predictions, check this checkbox.
class_sampling_factors: (Applicable only for classification and when balance_classes is enabled) Specify the per-class (in lexicographical order) over/under-sampling ratios. By default, these ratios are automatically computed during training to obtain the class balance.
max_after_balance_size: Specify the maximum relative size of the training data after balancing class counts (balance_classes must be enabled). The value can be less than 1.0.
overwrite_with_best_model: Check this checkbox to overwrite the final model with the best model found during training, based on the option selected for stopping_metric. This option is selected by default.
target_ratio_comm_to_comp: Specify the target ratio of communication overhead to computation. This option is only enabled for multi-node operation and if train_samples_per_iteration equals -2 (auto-tuning).
seed: Specify the random number generator (RNG) seed for algorithm components dependent on randomization. The seed is consistent for each H2O instance so that you can create models with the same starting conditions in alternative configurations.
rho: (Applicable only if adaptive_rate is enabled) Specify the adaptive learning rate time decay factor.
epsilon:(Applicable only if adaptive_rate is enabled) Specify the adaptive learning rate time smoothing factor to avoid dividing by zero.
max_w2: Specify the constraint for the squared sum of the incoming weights per unit (e.g., for Rectifier).
initial_weight_distribution: Select the initial weight distribution (Uniform Adaptive, Uniform, or Normal).
regression_stop: (Regression models only) Specify the stopping criterion for regression error (MSE) on the training data. To disable this option, enter -1.
diagnostics: Check this checkbox to compute the variable importances for input features (using the Gedeon method). For large networks, selecting this option can reduce speed. This option is selected by default.
fast_mode: Check this checkbox to enable fast mode, a minor approximation in back-propagation. This option is selected by default.
force_load_balance: Check this checkbox to force extra load balancing to increase training speed for small datasets and use all cores. This option is selected by default.
single_node_mode: Check this checkbox to force H2O to run on a single node for fine-tuning of model parameters. This option is not selected by default.
shuffle_training_data: Check this checkbox to shuffle the training data. This option is recommended if the training data is replicated and the value of train_samples_per_iteration is close to the number of nodes times the number of rows. This option is not selected by default.
missing_values_handling: Select how to handle missing values (skip or mean imputation).
quiet_mode: Check this checkbox to display less output in the standard output. This option is not selected by default.
sparse: Check this checkbox to enable sparse data handling, which is more efficient for data with many zero values.
col_major: Check this checkbox to use a column major weight matrix for the input layer. This option can speed up forward propagation but may reduce the speed of backpropagation. This option is not selected by default.
average_activation: Specify the average activation for the sparse autoencoder.
- If Rectifier is used, the average_activation value must be positive.
sparsity_beta: (Applicable only if autoencoder is enabled) Specify the sparsity-based regularization optimization. For more information, refer to the following link.
max_categorical_features: Specify the maximum number of categorical features enforced via hashing. The value must be at least one.
reproducible: To force reproducibility on small data, check this checkbox. If this option is enabled, the model takes more time to generate, since it uses only one thread.
export_weights_and_biases: To export the neural network weights and biases as H2O frames, check this checkbox.
elastic_averaging: To enable elastic averaging between computing nodes, which can improve distributed model convergence, check this checkbox (experimental).
rate: (Applicable only if adaptive_rate is disabled) Specify the learning rate. Higher values result in a less stable model, while lower values lead to slower convergence.
rate_annealing: (Applicable only if adaptive_rate is disabled) Specify the rate annealing value. The rate annealing is calculated as rate(1 + rate_annealing * samples).
rate_decay: (Applicable only if adaptive_rate is disabled) Specify the rate decay factor between layers. The rate decay is calculated as (N-th layer: rate * alpha^(N-1)).
momentum_start: (Applicable only if adaptive_rate is disabled) Specify the initial momentum at the beginning of training; we suggest 0.5.
momentum_ramp: (Applicable only if adaptive_rate is disabled) Specify the number of training samples for which the momentum increases.
momentum_stable: (Applicable only if adaptive_rate is disabled) Specify the final momentum after the ramp is over; we suggest 0.99.
nesterov_accelerated_gradient: (Applicable only if adaptive_rate is disabled) Enables the Nesterov Accelerated Gradient.
- initial_weight_scale: (Applicable only if initial_weight_distribution is Uniform or Normal) Specify the scale of the distribution function. For Uniform, the values are drawn uniformly. For Normal, the values are drawn from a Normal distribution with a standard deviation.
Interpreting a Deep Learning Model
To view the results, click the View button. The output for the Deep Learning model includes the following information for both the training and testing sets:
- Model parameters (hidden)
- A chart of the variable importances
- A graph of the scoring history (training MSE and validation MSE vs epochs)
- Output (model category, weights, biases)
- Status of neuron layers (layer number, units, type, dropout, L1, L2, mean rate, rate RMS, momentum, mean weight, weight RMS, mean bias, bias RMS)
- Scoring history in tabular format
- Training metrics (model name, model checksum name, frame name, frame checksum name, description, model category, duration in ms, scoring time, predictions, MSE, R2, logloss)
- Top-K Hit Ratios (for multi-class classification)
- Confusion matrix (for classification)
FAQ
- How does the algorithm handle missing values during training?
Deep Learning performs mean-imputation for missing numericals and creates a separate factor level for missing categoricals by default.
How does the algorithm handle missing values during testing?
Missing values in the test set will be mean-imputed during scoring.
What happens if the response has missing values?
No errors will occur, but nothing will be learned from rows containing missing the response.
Does it matter if the data is sorted?
Yes, since the training set is processed in order. Depending whether
train_samples_per_iterationis enabled, some rows will be skipped. Ifshuffle_training_datais enabled, then each thread that is processing a small subset of rows will process rows randomly, but it is not a global shuffle.Should data be shuffled before training?
Yes, the data should be shuffled before training, especially if the dataset is sorted.
How does the algorithm handle highly imbalanced data in a response column?
Specify
balance_classes,class_sampling_factorsandmax_after_balance_sizeto control over/under-sampling.What if there are a large number of columns?
The input neuron layer’s size is scaled to the number of input features, so as the number of columns increases, the model complexity increases as well.
What if there are a large number of categorical factor levels?
This is something to look out for. Say you have three columns: zip code (70k levels), height, and income. The resulting number of internally one-hot encoded features will be 70,002 and only 3 of them will be activated (non-zero). If the first hidden layer has 200 neurons, then the resulting weight matrix will be of size 70,002 x 200, which can take a long time to train and converge. In this case, we recommend either reducing the number of categorical factor levels upfront (e.g., using
h2o.interaction()from R), or specifyingmax_categorical_featuresto use feature hashing to reduce the dimensionality.How does your Deep Learning Autoencoder work? Is it deep or shallow?
H2O’s DL autoencoder is based on the standard deep (multi-layer) neural net architecture, where the entire network is learned together, instead of being stacked layer-by-layer. The only difference is that no response is required in the input and that the output layer has as many neurons as the input layer. If you don’t achieve convergence, then try using the Tanh activation and fewer layers. We have some example test scripts here, and even some that show how stacked auto-encoders can be implemented in R.
When building the model, does Deep Learning use all features or a selection of the best features?
For Deep Learning, all features are used, unless you manually specify that columns should be ignored. Adding an L1 penalty can make the model sparse, but it is still the full size.
What is the relationship between iterations, epochs, and the
train_samples_per_iterationparameter?Epochs measures the amount of training. An iteration is one MapReduce (MR) step - essentially, one pass over the data. The
train_samples_per_iterationparameter is the amount of data to use for training for each MR step, which can be more or less than the number of rows.
When do
reduce()calls occur, after each iteration or each epoch?Neither;
reduce()calls occur after every twomap()calls, between threads and ultimately between nodes. There are manyreduce()calls, much more than one per MapReduce step (also known as an “iteration”). Epochs are not related to MR iterations, unless you specifytrain_samples_per_iterationas0or-1(or to number of rows/nodes). Otherwise, one MR iteration can train with an arbitrary number of training samples (as specified bytrain_samples_per_iteration).Does each Mapper task work on a separate neural-net model that is combined during reduction, or is each Mapper manipulating a shared object that’s persistent across nodes?
Neither; there’s one model per compute node, so multiple Mappers/threads share one model, which is why H2O is not reproducible unless a small dataset is used and
force_load_balance=Forreproducible=T, which effectively rebalances to a single chunk and leads to only one thread to launch amap(). The current behavior is simple model averaging; between-node model averaging via “Elastic Averaging” is currently in progress.Is the loss function and backpropagation performed after each individual training sample, each iteration, or at the epoch level?
Loss function and backpropagation are performed after each training sample (mini-batch size 1 == online stochastic gradient descent).
When using Hinton’s dropout and specifying an input dropout ratio of ~20% and
train_samples_per_iterationis set to 50, will each of the 50 samples have a different set of the 20% input neurons suppressed?Yes - suppression is not done at the iteration level across as samples in that iteration. The dropout mask is different for each training sample.
When using dropout parameters such as
input_dropout_ratio, what happens if you use onlyRectifierinstead ofRectifierWithDropoutin the activation parameter?The amount of dropout on the input layer can be specified for all activation functions, but hidden layer dropout is only supported is set to
WithDropout. The default hidden dropout is 50%, so you don’t need to specify anything but the activation type to get good results, but you can set the hidden dropout values for each layer separately.When using the
score_validation_samplingandscore_training_samplesparameters, is scoring done at the end of the Deep Learning run?The majority of scoring takes place after each MR iteration. After the iteration is complete, it may or may not be scored, depending on two criteria: the time since the last scoring and the time needed for scoring.
The maximum time between scoring (
score_interval, default = 5 seconds) and the maximum fraction of time spent scoring (score_duty_cycle) independently of loss function, backpropagation, etc.Of course, using more training or validation samples will increase the time for scoring, as well as scoring more frequently. For more information about how this affects runtime, refer to the Deep Learning Performance Guide.
How does the validation frame affect the built neuron network?
The validation frame is only used for scoring and does not directly affect the model. However, the validation frame can be used stopping the model early if
overwrite_with_best_model = T, which is the default. If this parameter is enabled, the model with the lowest validation error is displayed at the end of the training.By default, the validation frame is used to tune the model parameters (such as number of epochs) and will return the best model as measured by the validation metrics, depending on how often the validation metrics are computed (
score_duty_cycle) and whether the validation frame itself was sampled.Model-internal sampling of the validation frame (
score_validation_samplesandscore_validation_samplingfor optional stratification) will affect early stopping quality. If you specify a validation frame but setscore_validation_samplesto more than the number of rows in the validation frame (instead of 0, which represents the entire frame), the validation metrics received at the end of training will not be reproducible, since the model does internal sampling.Are there any best practices for building a model using checkpointing?
In general, to get the best possible model, we recommend building a model with train\_samples\_per\_iteration = -2 (which is the default value for auto-tuning) and saving it.
To improve the initial model, start from the previous model and add iterations by building another model, setting the checkpoint to the previous model, and changing train\_samples\_per\_iteration, target\_ratio\_comm\_to\_comp, or other parameters.
If you don’t know your model ID because it was generated by R, look it up using h2o.ls(). By default, Deep Learning model names start with deeplearning_ To view the model, use m <- h2o.getModel("my\_model\_id") or summary(m).
There are a few ways to manage checkpoint restarts:
Option 1: (Multi-node only) Leave train\_samples\_per\_iteration = -2, increase target\_comm\_to\_comp from 0.05 to 0.25 or 0.5, which provides more communication. This should result in a better model when using multiple nodes. Note: This does not affect single-node performance.
Option 2: (Single or multi-node) Set train\_samples\_per\_iteration to \(N\), where \(N\) is the number of training samples used for training by the entire cluster for one iteration. Each of the nodes then trains on \(N\) randomly-chosen rows for every iteration. The number defined as \(N\) depends on the dataset size and the model complexity.
Option 3: (Single or multi-node) Change regularization parameters such as l1, l2, max\_w2, input\_droput\_ratio or hidden\_dropout\_ratios. We recommend build the first mode using RectifierWithDropout, input\_dropout\_ratio = 0 (if there is suspected noise in the input), and hidden\_dropout\_ratios=c(0,0,0) (for the ability to enable dropout regularization later).
- How does class balancing work?
The max\_after\_balance\_size parameter defines the maximum size of the over-sampled dataset. For example, if max\_after\_balance\_size = 3, the over-sampled dataset will not be greater than three times the size of the original dataset.
For example, if you have five classes with priors of 90%, 2.5%, 2.5%, and 2.5% (out of a total of one million rows) and you oversample to obtain a class balance using balance\_classes = T, the result is all four minor classes are oversampled by forty times and the total dataset will be 4.5 times as large as the original dataset (900,000 rows of each class). If max\_after\_balance\_size = 3, all five balance classes are reduced by 3/5 resulting in 600,000 rows each (three million total).
To specify the per-class over- or under-sampling factors, use class\_sampling\_factors. In the previous example, the default behavior with balance\_classes is equivalent to c(1,40,40,40,40), while when max\_after\_balance\_size = 3, the results would be c(3/5,40*3/5,40*3/5,40*3/5).
In all cases, the probabilities are adjusted to the pre-sampled space, so the minority classes will have lower average final probabilities than the majority class, even if they were sampled to reach class balance.
- How is variable importance calculated for Deep Learning?
For Deep Learning, variable importance is calculated using the Gedeon method.
Deep Learning Algorithm
To compute deviance for a Deep Learning regression model, the following formula is used:
Loss = Quadratic -> MSE==Deviance For Absolute/Laplace or Huber -> MSE != Deviance
For more information about how the Deep Learning algorithm works, refer to the Deep Learning booklet.
References
Candel, Arno and Parmar, Viraj. “Deep Learning with H2O.” H2O.ai, Inc. (2015).
Candel, Arno. “The Definitive Performance Tuning Guide for H2O Deep Learning.” H2O.ai, Inc. (2015).
Cross-Validation
N-fold cross-validation is used to validate a model internally, i.e., estimate the model performance without having to sacrifice a validation split. Also, you avoid statistical issues with your validation split (it might be a “lucky” split, especially for imbalanced data). Good values for N are around 5 to 10. Comparing the N validation metrics is always a good idea, to check the stability of the estimation, before “trusting” the main model.
You have to make sure, however, that the holdout sets for each of the N models are good. For i.i.d. data, the random splitting of the data into N pieces (default behavior) or modulo-based splitting is fine. For temporal or otherwise structured data with distinct “events”, you have to make sure to split the folds based on the events. For example, if you have observations (e.g., user transactions) from N cities and you want to build models on users from only N-1 cities and validate them on the remaining city (if you want to study the generalization to new cities, for example), you will need to specify the parameter “fold_column” to be the city column. Otherwise, you will have rows (users) from all N cities randomly blended into the N folds, and all N cv models will see all N cities, making the validation less useful (or totally wrong, depending on the distribution of the data). This is known as “data leakage”: https://youtu.be/NHw_aKO5KUM?t=889
How Cross-Validation is Calculated
In general, for all algos that support the nfolds parameter, H2O’s cross-validation works as follows:
For example, for nfolds=5, 6 models are built. The first 5 models (cross-validation models) are built on 80% of the training data, and a different 20% is held out for each of the 5 models. Then the main model is built on 100% of the training data. This main model is the model you get back from H2O in R, Python and Flow.
This main model contains training metrics and cross-validation metrics (and optionally, validation metrics if a validation frame was provided). The main model also contains pointers to the 5 cross-validation models for further inspection.
All 5 cross-validation models contain training metrics (from the 80% training data) and validation metrics (from their 20% holdout/validation data). To compute their individual validation metrics, each of the 5 cross-validation models had to make predictions on their 20% of of rows of the original training frame, and score against the true labels of the 20% holdout.
For the main model, this is how the cross-validation metrics are computed: The 5 holdout predictions are combined into one prediction for the full training dataset (i.e., predictions for every row of the training data, but the model making the prediction for a particular row has not seen that row during training). This “holdout prediction” is then scored against the true labels, and the overall cross-validation metrics are computed.
This approach has some implications. Scoring the holdout predictions freshly can result in different metrics than taking the average of the 5 validation metrics of the cross-validation models. For example, if the sizes of the holdout folds differ a lot (e.g., when a user-given fold_column is used), then the average should probably be replaced with a weighted average. Also, if the cross-validation models map to slightly different probability spaces, which can happen for small DL models that converge to different local minima, then the confused rank ordering of the combined predictions would lead to a significantly different AUC than the average.
Example in R
To gain more insights into the variance of the holdout metrics (e.g., AUCs), you can look up the cross-validation models, and inspect their validation metrics. Here’s an R code example showing the two approaches:
library(h2o)
h2o.init()
df <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/prostate/prostate.csv.zip")
df$CAPSULE <- as.factor(df$CAPSULE)
model_fit <- h2o.gbm(3:8,2,df,nfolds=5,seed=1234)
# Default: AUC of holdout predictions
h2o.auc(model_fit,xval=TRUE)
# Optional: Average the holdout AUCs
cvAUCs <- sapply(sapply(model_fit@model$cross_validation_models, `[[`, "name"), function(x) { h2o.auc(h2o.getModel(x), valid=TRUE) })
print(cvAUCs)
mean(cvAUCs)
Using Cross-Validated Predictions
With cross-validated model building, H2O builds N+1 models: N cross-validated model and 1 overarching model over all of the training data.
Each cv-model produces a prediction frame pertaining to its fold. It can be saved and probed from the various clients if keep_cross_validation_predictions parameter is set in the model constructor.
These holdout predictions have some interesting properties. First they have names like:
prediction_GBM_model_1452035702801_1_cv_1
and they contain, unsurprisingly, predictions for the data held out in the fold. They also have the same number of rows as the entire input training frame with 0s filled in for all rows that are not in the hold out.
Let’s look at an example.
Here is a snippet of a three-class classification dataset (last column is the response column), with a 3-fold identification column appended to the end:
| sepal_len | sepal_wid | petal_len | petal_wid | class | foldId |
|---|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa | 0 |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa | 0 |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa | 2 |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa | 1 |
| 5.0 | 3.6 | 1.4 | 0.2 | setosa | 2 |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa | 1 |
| 4.6 | 3.4 | 1.4 | 0.3 | setosa | 1 |
| 5.0 | 3.4 | 1.5 | 0.2 | setosa | 0 |
| 4.4 | 2.9 | 1.4 | 0.4 | setosa | 1 |
Each cross-validated model produces a prediction frame
prediction_GBM_model_1452035702801_1_cv_1
prediction_GBM_model_1452035702801_1_cv_2
prediction_GBM_model_1452035702801_1_cv_3
and each one has the following shape (for example the first one):
prediction_GBM_model_1452035702801_1_cv_1
| prediction | setosa | versicolor | virginica |
|---|---|---|---|
| 1 | 0.0232 | 0.7321 | 0.2447 |
| 2 | 0.0543 | 0.2343 | 0.7114 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0 | 0 | 0 |
| 0 | 0.8921 | 0.0321 | 0.0758 |
| 0 | 0 | 0 | 0 |
The training rows receive a prediction of 0 (more on this below) as well as 0 for all class probabilities. Each of these holdout predictions has the same number of rows as the input frame.
Combining holdout predictions
The frame of cross-validated predictions is simply the superposition of the individual predictions. Here’s an example from R:
library(h2o)
h2o.init()
# H2O Cross-validated K-means example
prosPath <- system.file("extdata", "prostate.csv", package="h2o")
prostate.hex <- h2o.uploadFile(path = prosPath)
fit <- h2o.kmeans(training_frame = prostate.hex,
k = 10,
x = c("AGE", "RACE", "VOL", "GLEASON"),
nfolds = 5, #If you want to specify folds directly, then use "fold_column" arg
keep_cross_validation_predictions = TRUE)
# This is where cv preds are stored:
fit@model$cross_validation_predictions$name
# Compress the CV preds into a single H2O Frame:
# Each fold's preds are stored in a N x 1 col, where the row values for non-active folds are set to zero
# So we will compress this into a single 1-col H2O Frame (easier to digest)
nfolds <- fit@parameters$nfolds
predlist <- sapply(1:nfolds, function(v) h2o.getFrame(fit@model$cross_validation_predictions[[v]]$name)$predict, simplify = FALSE)
cvpred_sparse <- h2o.cbind(predlist) # N x V Hdf with rows that are all zeros, except corresponding to the v^th fold if that rows is associated with v
pred <- apply(cvpred_sparse, 1, sum) # These are the cross-validated predicted cluster IDs for each of the 1:N observations
This can be extended to other family types as well (multinomial, binomial, regression):
# helper function
.compress_to_cvpreds <- function(h2omodel, family) {
# return the frame_id of the resulting 1-col Hdf of cvpreds for learner l
V <- h2omodel@allparameters$nfolds
if (family %in% c("bernoulli", "binomial")) {
predlist <- sapply(1:V, function(v) h2o.getFrame(h2omodel@model$cross_validation_predictions[[v]]$name)[,3], simplify = FALSE)
} else {
predlist <- sapply(1:V, function(v) h2o.getFrame(h2omodel@model$cross_validation_predictions[[v]]$name)$predict, simplify = FALSE)
}
cvpred_sparse <- h2o.cbind(predlist) # N x V Hdf with rows that are all zeros, except corresponding to the v^th fold if that rows is associated with v
cvpred_col <- apply(cvpred_sparse, 1, sum)
return(cvpred_col)
}
# Extract cross-validated predicted values (in order of original rows)
h2o.cvpreds <- function(object) {
# Need to extract family from model object
if (class(object) == "H2OBinomialModel") family <- "binomial"
if (class(object) == "H2OMulticlassModel") family <- "multinomial"
if (class(object) == "H2ORegressionModel") family <- "gaussian"
cvpreds <- .compress_to_cvpreds(h2omodel = object, family = family)
return(cvpreds)
}
YARN Best Practices
- Using H2O with YARN
- Configuring YARN
- Limiting CPU Usage
- Specifying Queues
- Specifying Output Directories
- Customizing YARN
- Accessing Logs
YARN (Yet Another Resource Manager) is a resource management framework. H2O can be launched as an application on YARN. If you want to run H2O on Hadoop, essentially, you are running H2O on YARN. If you are not currently using YARN to manage your cluster resources, we strongly recommend it.
Using H2O with YARN
When you launch H2O on Hadoop using the hadoop jar command, YARN allocates the necessary resources to launch the requested number of nodes. H2O launches as a MapReduce (V2) task, where each mapper is an H2O node of the specified size.
hadoop jar h2odriver.jar -nodes 1 -mapperXmx 6g -output hdfsOutputDirName
Occasionally, YARN may reject a job request. This usually occurs because either there is not enough memory to launch the job or because of an incorrect configuration.
If YARN rejects the job request, try launching the job with less memory to see if that is the cause of the failure. Specify smaller values for -mapperXmx (we recommend a minimum of 2g) and -nodes (start with 1) to confirm that H2O can launch successfully.
To resolve configuration issues, adjust the maximum memory that YARN will allow when launching each mapper. If the cluster manager settings are configured for the default maximum memory size but the memory required for the request exceeds that amount, YARN will not launch and H2O will time out. If you are using the default configuration, change the configuration settings in your cluster manager to specify memory allocation when launching mapper tasks. To calculate the amount of memory required for a successful launch, use the following formula:
YARN container size (
mapreduce.map.memory.mb) =-mapperXmxvalue + (-mapperXmx*-extramempercent[default is 10%])
The mapreduce.map.memory.mb value must be less than the YARN memory configuration values for the launch to succeed.
Configuring YARN
For Cloudera, configure the settings in Cloudera Manager. Depending on how the cluster is configured, you may need to change the settings for more than one role group.
Click Configuration and enter the following search term in quotes: yarn.nodemanager.resource.memory-mb.
Enter the amount of memory (in GB) to allocate in the Value field. If more than one group is listed, change the values for all listed groups.
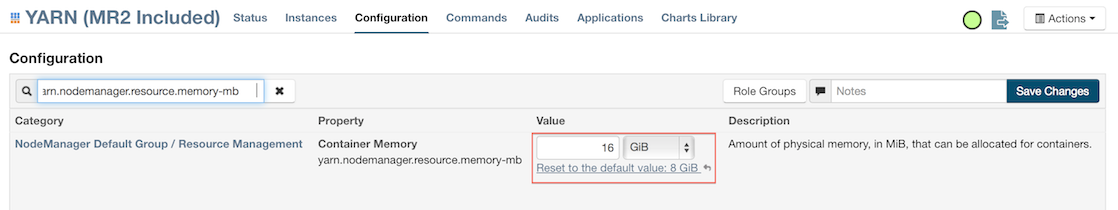
Click the Save Changes button in the upper-right corner.
- Enter the following search term in quotes: yarn.scheduler.maximum-allocation-mb
Change the value, click the Save Changes button in the upper-right corner, and redeploy.
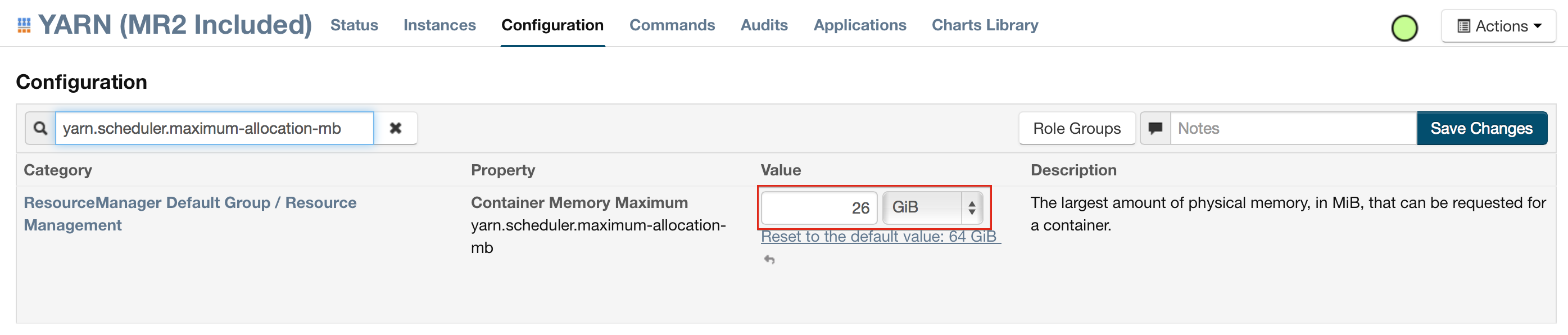
For Hortonworks, configure the settings in Ambari.
- Select YARN, then click the Configs tab.
- Select the group.
In the Node Manager section, enter the amount of memory (in MB) to allocate in the yarn.nodemanager.resource.memory-mb entry field.
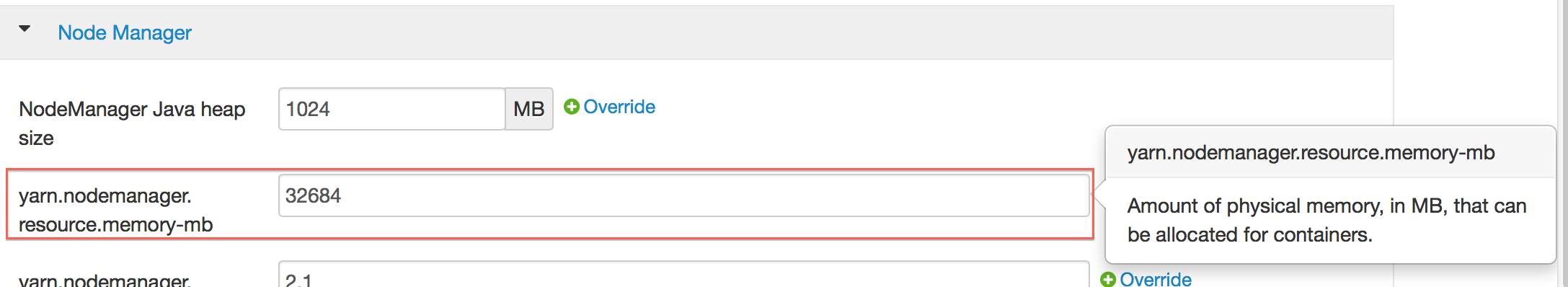
In the Scheduler section, enter the amount of memory (in MB)to allocate in the yarn.scheduler.maximum-allocation-mb entry field.
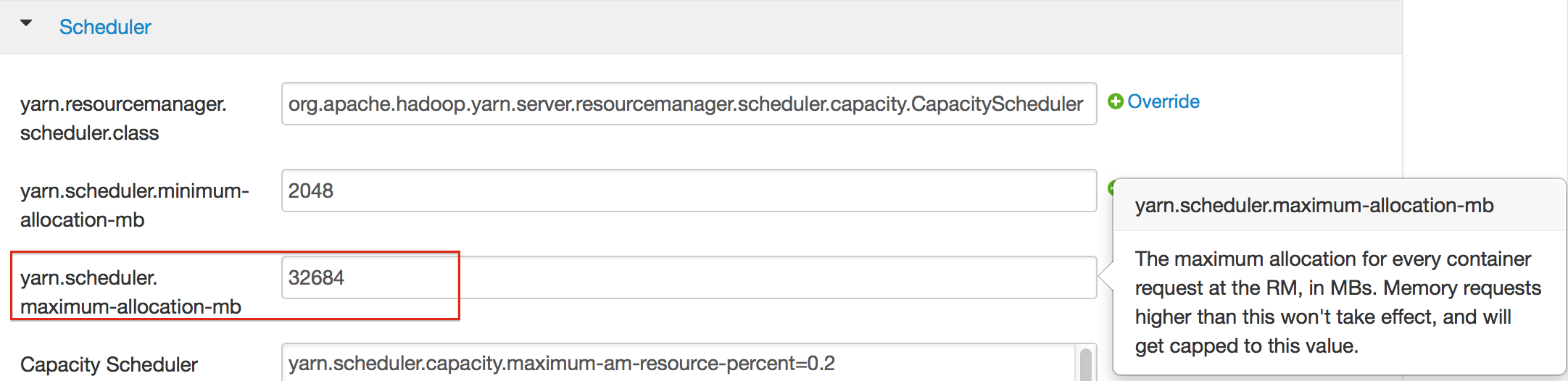
Click the Save button at the bottom of the page and redeploy the cluster.
For MapR:
- Edit the yarn-site.xml file for the node running the ResourceManager.
- Change the values for the
yarn.nodemanager.resource.memory-mbandyarn.scheduler.maximum-allocation-mbproperties. - Restart the ResourceManager and redeploy the cluster.
To verify the values were changed, check the values for the following properties:
- <name>yarn.nodemanager.resource.memory-mb</name>
- <name>yarn.scheduler.maximum-allocation-mb</name>
Limiting CPU Usage
To limit the number of CPUs used by H2O, use the -nthreads option and specify the maximum number of CPUs for a single container to use. The following example limits the number of CPUs to four:
hadoop jar h2odriver.jar -nthreads 4 -nodes 1 -mapperXmx 6g -output hdfsOutputDirName
Note: The default is 4*the number of CPUs. You must specify at least four CPUs; otherwise, the following error message displays:
ERROR: nthreads invalid (must be >= 4)
Specifying Queues
If you do not specify a queue when launching H2O, H2O jobs are submitted to the default queue. Jobs submitted to the default queue have a lower priority than jobs submitted to a specific queue.
To specify a queue with Hadoop, enter -Dmapreduce.job.queuename=<my-h2o-queue>
(where <my-h2o-queue> is the name of the queue) when launching Hadoop.
For example,
hadoop jar h2odriver.jar -Dmapreduce.job.queuename=<my-h2o-queue> -nodes <num-nodes> -mapperXmx 6g -output hdfsOutputDirName
Specifying Output Directories
To prevent overwriting multiple users’ files, each job must have a unique output directory name. Change the -output hdfsOutputDir argument (where hdfsOutputDir is the name of the directory.
Alternatively, you can delete the directory (manually or by using a script) instead of creating a unique directory each time you launch H2O.
Customizing YARN
Most of the configurable YARN variables are stored in yarn-site.xml. To prevent settings from being overridden, you can mark a config as “final.” If you change any values in yarn-site.xml, you must restart YARN to confirm the changes.
Accessing Logs
To learn how to access logs in YARN, refer to Downloading Logs.
Downloading Logs
Accessing Logs
Depending on whether you are using Hadoop with H2O and whether the job is currently running, there are different ways of obtaining the logs for H2O.
Copy and email the logs to support@h2o.ai or submit them to h2ostream@googlegroups.com with a brief description of your Hadoop environment, including the Hadoop distribution and version.
Without Running Jobs
- If you are using Hadoop and the job is not running, view the logs by using the
yarn logs -applicationIdcommand. When you start an H2O instance, the complete command displays in the output:
jessica@mr-0x8:~/h2o-3.1.0.3008-cdh5.2$ hadoop jar h2odriver.jar -nodes 1 -mapperXmx 6g -output hdfsOutputDirName
Determining driver host interface for mapper->driver callback...
[Possible callback IP address: 172.16.2.178]
[Possible callback IP address: 127.0.0.1]
Using mapper->driver callback IP address and port: 172.16.2.178:52030
(You can override these with -driverif and -driverport.)
Memory Settings:
mapreduce.map.java.opts: -Xms1g -Xmx1g -Dlog4j.defaultInitOverride=true
Extra memory percent: 10
mapreduce.map.memory.mb: 1126
15/05/06 17:11:50 INFO client.RMProxy: Connecting to ResourceManager at mr-0x10.0xdata.loc/172.16.2.180:8032
15/05/06 17:11:52 INFO mapreduce.JobSubmitter: number of splits:1
15/05/06 17:11:52 INFO mapreduce.JobSubmitter: Submitting tokens for job: job_1430127035640_0075
15/05/06 17:11:52 INFO impl.YarnClientImpl: Submitted application application_1430127035640_0075
15/05/06 17:11:52 INFO mapreduce.Job: The url to track the job: http://mr-0x10.0xdata.loc:8088/proxy/application_1430127035640_0075/
Job name 'H2O_29570' submitted
JobTracker job ID is 'job_1430127035640_0075'
For YARN users, logs command is 'yarn logs -applicationId application_1430127035640_0075'
Waiting for H2O cluster to come up...
In the above example, the command is specified in the next to last line (For YARN users, logs command is...). The command is unique for each instance. In Terminal, enter yarn logs -applicationId application_<UniqueID> to view the logs (where <UniqueID> is the number specified in the next to last line of the output that displayed when you created the cluster).
Use YARN to obtain the stdout and stderr logs that are used for troubleshooting. To learn how to access YARN based on management software, version, and job status, see Accessing YARN.
Click the Applications link to view all jobs, then click the History link for the job.
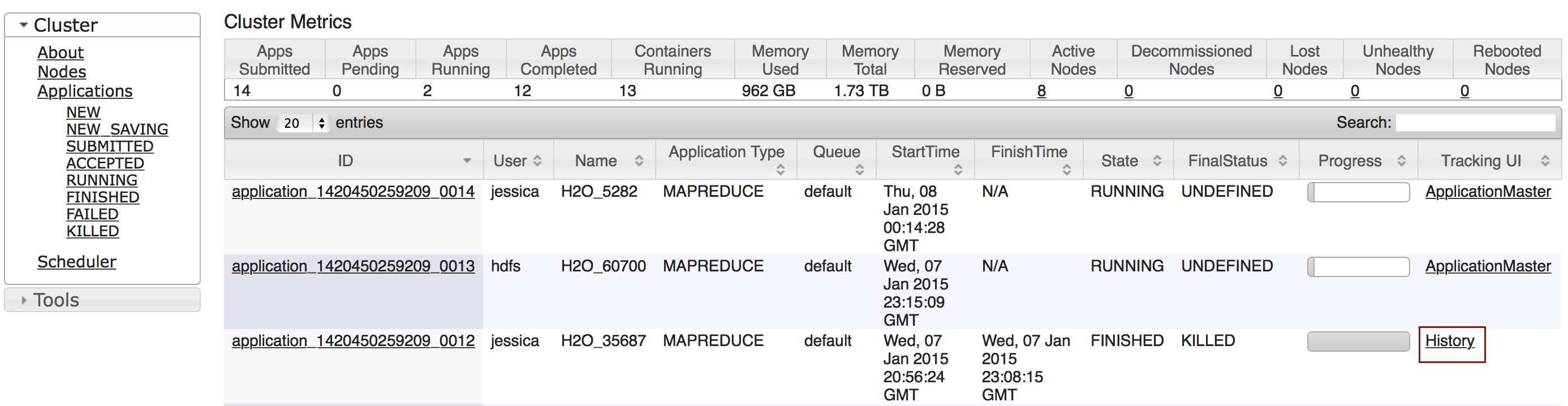
Click the logs link.
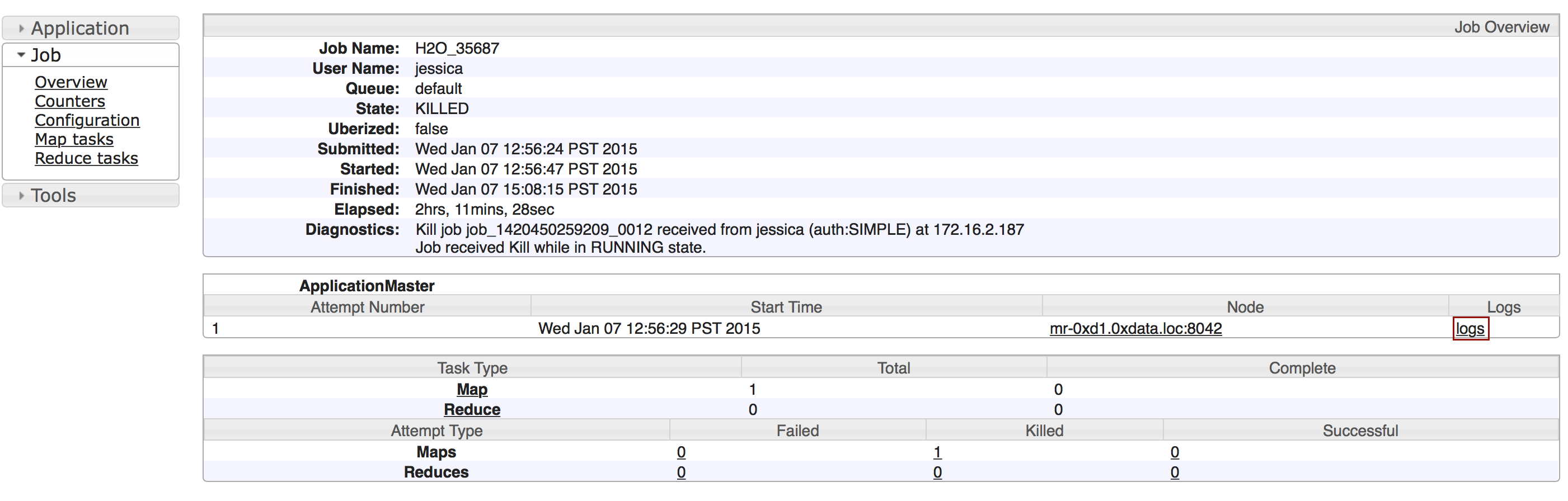
Copy the information that displays and send it in an email to support@h2o.ai.
With Running Jobs
If you are using Hadoop and the job is still running:
Use YARN to obtain the
stdoutandstderrlogs that are used for troubleshooting. To learn how to access YARN based on management software, version, and job status, see Accessing YARN.Click the Applications link to view all jobs, then click the ApplicationMaster link for the job.
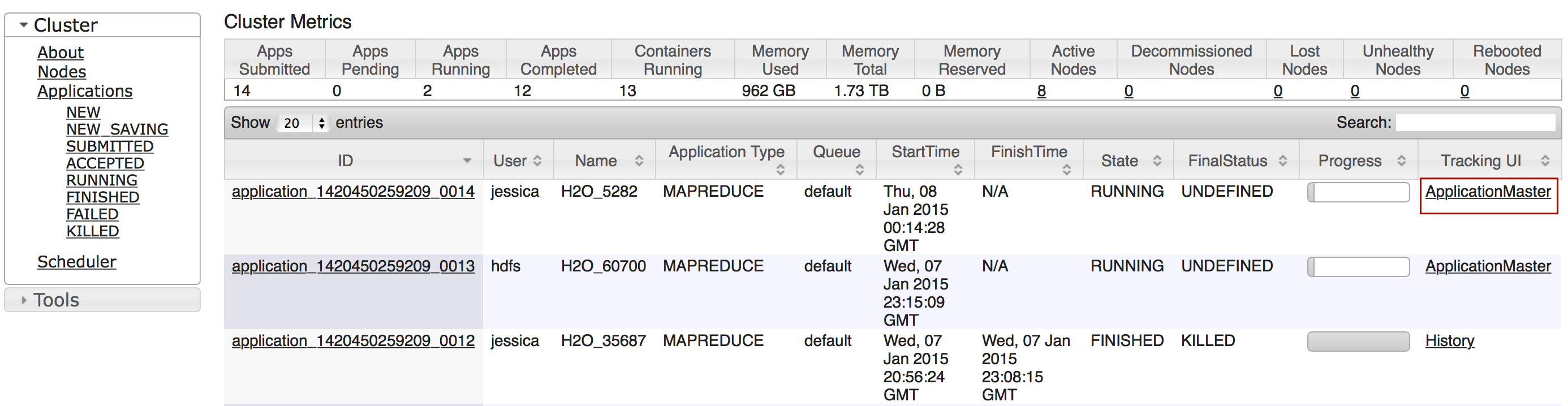
Select the job from the list of active jobs.
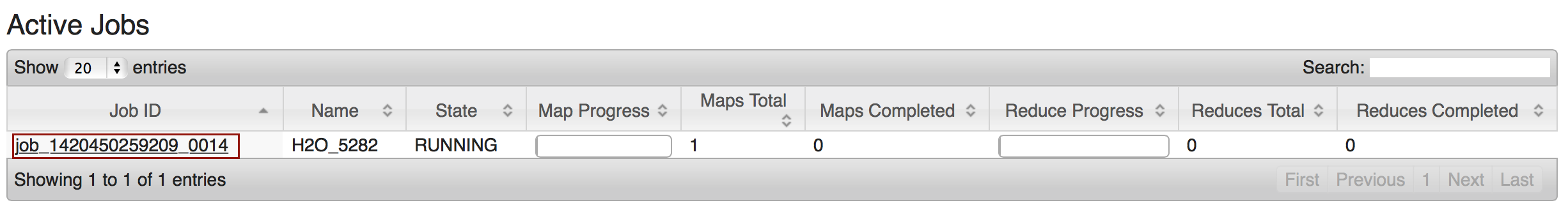
Click the logs link.
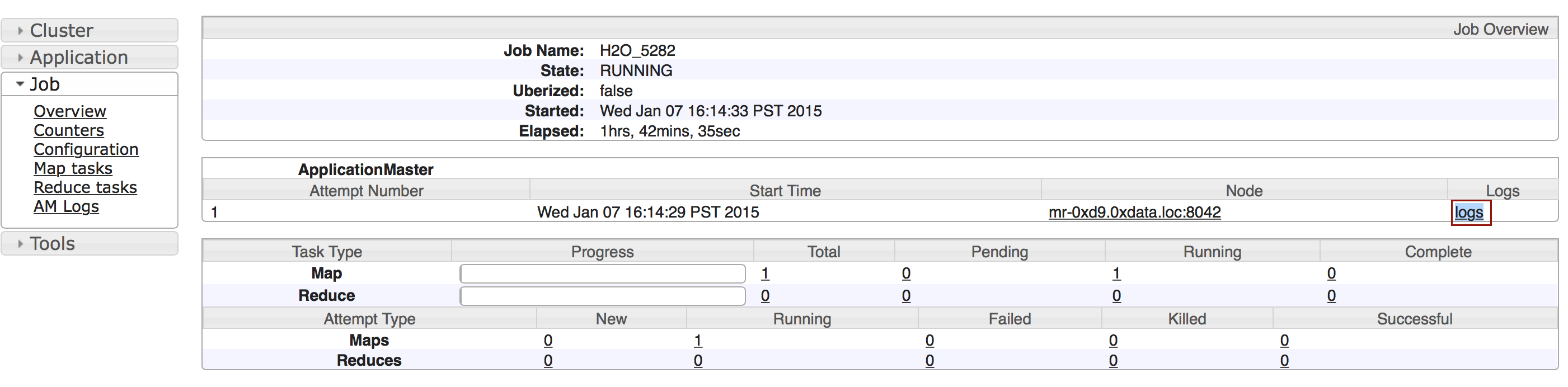
Send the contents of the displayed files to support@h2o.ai.

Go to the H2O web UI and select Admin > View Log. To filter the results select a node or log file type from the drop-down menus. To download the logs, click the Download Logs button.
When you view the log, the output displays the location of log directory after
Log dir:(as shown in the last line in the following example):
05-06 17:12:15.610 172.16.2.179:54321 26336 main INFO: ----- H2O started -----
05-06 17:12:15.731 172.16.2.179:54321 26336 main INFO: Build git branch: master
05-06 17:12:15.731 172.16.2.179:54321 26336 main INFO: Build git hash: 41d039196088df081ad77610d3e2d6550868f11b
05-06 17:12:15.731 172.16.2.179:54321 26336 main INFO: Build git describe: jenkins-master-1187
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Build project version: 0.3.0.1187
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Built by: 'jenkins'
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Built on: '2015-05-05 23:31:12'
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java availableProcessors: 8
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java heap totalMemory: 982.0 MB
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java heap maxMemory: 982.0 MB
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java version: Java 1.7.0_80 (from Oracle Corporation)
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: OS version: Linux 3.13.0-51-generic (amd64)
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Machine physical memory: 31.30 GB
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: X-h2o-cluster-id: 1430957535344
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Possible IP Address: virbr0 (virbr0), 192.168.122.1
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Possible IP Address: br0 (br0), 172.16.2.179
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Possible IP Address: lo (lo), 127.0.0.1
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Multiple local IPs detected:
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: /192.168.122.1 /172.16.2.179
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Attempting to determine correct address...
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Using /172.16.2.179
05-06 17:12:15.734 172.16.2.179:54321 26336 main INFO: Internal communication uses port: 54322
05-06 17:12:15.734 172.16.2.179:54321 26336 main INFO: Listening for HTTP and REST traffic on http://172.16.2.179:54321/
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: H2O cloud name: 'H2O_29570' on /172.16.2.179:54321, discovery address /237.61.246.13:60733
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: If you have trouble connecting, try SSH tunneling from your local machine (e.g., via port 55555):
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: 1. Open a terminal and run 'ssh -L 55555:localhost:54321 yarn@172.16.2.179'
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: 2. Point your browser to http://localhost:55555
05-06 17:12:15.979 172.16.2.179:54321 26336 main INFO: Log dir: '/home2/yarn/nm/usercache/jessica/appcache/application_1430127035640_0075/h2ologs'
- In Terminal, enter
cd /tmp/h2o-<UserName>/h2ologs(where<UserName>is your computer user name), then enterls -lto view a list of the log files. Thehttpdlog contains the request/response status of all REST API transactions. The rest of the logs use the formath2o_\<IPaddress>\_<Port>-<LogLevel>-<LogLevelName>.log, where<IPaddress>is the bind address of the H2O instance,<Port>is the port number,<LogLevel>is the numerical log level (1-6, with 6 as the highest severity level), and<LogLevelName>is the name of the log level (trace, debug, info, warn, error, or fatal).
- Download the logs using R. In R, enter the command
h2o.downloadAllLogs(filename = "logs.zip")(wherefilenameis the specified filename for the logs).
Accessing YARN
Methods for accessing YARN vary depending on the default management software and version, as well as job status.
Cloudera 5 & 5.2
In Cloudera Manager, click the YARN link in the cluster section.
In the Quick Links section, select ResourceManager Web UI if the job is running or select HistoryServer Web UI if the job is not running.
Ambari
From the Ambari Dashboard, select YARN.
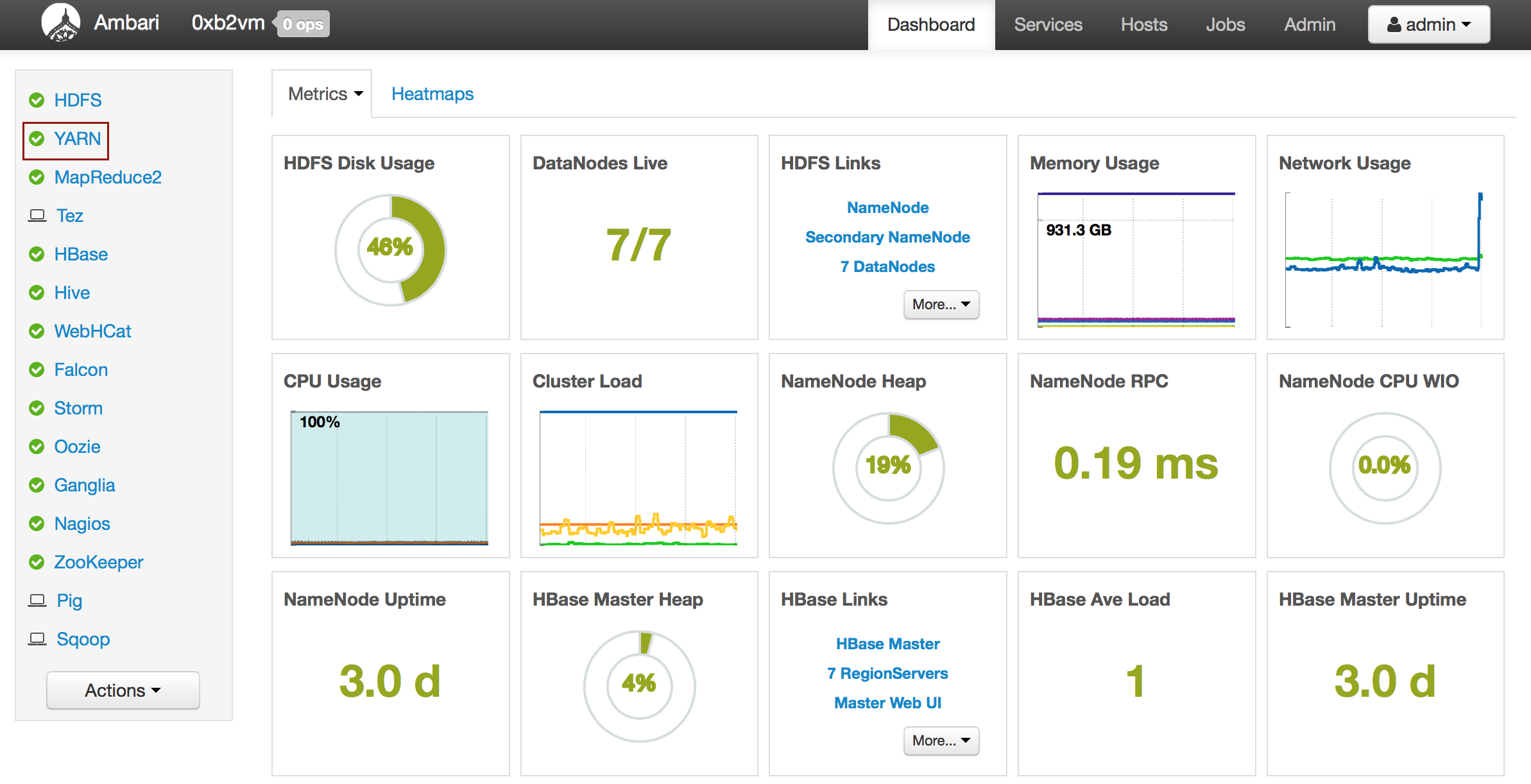
From the Quick Links drop-down menu, select ResourceManager UI.
For Non-Hadoop Users
Without Current Jobs
If you are not using Hadoop and the job is not running:
- In Terminal, enter
cd /tmp/h2o-<UserName>/h2ologs(where<UserName>is your computer user name), then enterls -lto view a list of the log files. Thehttpdlog contains the request/response status of all REST API transactions. The rest of the logs use the formath2o_\<IPaddress>\_<Port>-<LogLevel>-<LogLevelName>.log, where<IPaddress>is the bind address of the H2O instance,<Port>is the port number,<LogLevel>is the numerical log level (1-6, with 6 as the highest severity level), and<LogLevelName>is the name of the log level (trace, debug, info, warn, error, or fatal).
With Current Jobs
If you are not using Hadoop and the job is still running:
Go to the H2O web UI and select Admin > Inspect Log or go to http://localhost:54321/LogView.html.
To download the logs, click the Download Logs button.
When you view the log, the output displays the location of log directory after
Log dir:(as shown in the last line in the following example):
05-06 17:12:15.610 172.16.2.179:54321 26336 main INFO: ----- H2O started -----
05-06 17:12:15.731 172.16.2.179:54321 26336 main INFO: Build git branch: master
05-06 17:12:15.731 172.16.2.179:54321 26336 main INFO: Build git hash: 41d039196088df081ad77610d3e2d6550868f11b
05-06 17:12:15.731 172.16.2.179:54321 26336 main INFO: Build git describe: jenkins-master-1187
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Build project version: 0.3.0.1187
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Built by: 'jenkins'
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Built on: '2015-05-05 23:31:12'
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java availableProcessors: 8
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java heap totalMemory: 982.0 MB
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java heap maxMemory: 982.0 MB
05-06 17:12:15.732 172.16.2.179:54321 26336 main INFO: Java version: Java 1.7.0_80 (from Oracle Corporation)
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: OS version: Linux 3.13.0-51-generic (amd64)
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Machine physical memory: 31.30 GB
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: X-h2o-cluster-id: 1430957535344
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Possible IP Address: virbr0 (virbr0), 192.168.122.1
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Possible IP Address: br0 (br0), 172.16.2.179
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Possible IP Address: lo (lo), 127.0.0.1
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Multiple local IPs detected:
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: /192.168.122.1 /172.16.2.179
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Attempting to determine correct address...
05-06 17:12:15.733 172.16.2.179:54321 26336 main INFO: Using /172.16.2.179
05-06 17:12:15.734 172.16.2.179:54321 26336 main INFO: Internal communication uses port: 54322
05-06 17:12:15.734 172.16.2.179:54321 26336 main INFO: Listening for HTTP and REST traffic on http://172.16.2.179:54321/
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: H2O cloud name: 'H2O_29570' on /172.16.2.179:54321, discovery address /237.61.246.13:60733
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: If you have trouble connecting, try SSH tunneling from your local machine (e.g., via port 55555):
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: 1. Open a terminal and run 'ssh -L 55555:localhost:54321 yarn@172.16.2.179'
05-06 17:12:15.744 172.16.2.179:54321 26336 main INFO: 2. Point your browser to http://localhost:55555
05-06 17:12:15.979 172.16.2.179:54321 26336 main INFO: Log dir: '/home2/yarn/nm/usercache/jessica/appcache/application_1430127035640_0075/h2ologs'
- In Terminal, enter
cd /tmp/h2o-<UserName>/h2ologs(where<UserName>is your computer user name), then enterls -lto view a list of the log files. Thehttpdlog contains the request/response status of all REST API transactions. The rest of the logs use the formath2o_\<IPaddress>\_<Port>-<LogLevel>-<LogLevelName>.log, where<IPaddress>is the bind address of the H2O instance,<Port>is the port number,<LogLevel>is the numerical log level (1-6, with 6 as the highest severity level), and<LogLevelName>is the name of the log level (trace, debug, info, warn, error, or fatal).
To view the REST API logs from R:
In R, enter
h2o.startLogging(). The output displays the location of the REST API logs:> h2o.startLogging() Appending REST API transactions to log file /var/folders/ylcq5nhky53hjcl9wrqxt39kz80000gn/T//RtmpE7X8Yv/rest.logCopy the displayed file path. In Terminal, enter
lessand paste the file path.- Press Enter. A time-stamped log of all REST API transactions displays.
------------------------------------------------------------
Time: 2015-01-06 15:46:11.083
GET http://172.16.2.20:54321/3/Cloud.json
postBody:
curlError: FALSE
curlErrorMessage:
httpStatusCode: 200
httpStatusMessage: OK
millis: 3
{"__meta":{"schema_version": 1,"schema_name":"CloudV1","schema_type":"Iced"},"version":"0.1.17.1009","cloud_name":...[truncated]}
-------------------------------------------------------------
- Download the logs using R. In R, enter the command
h2o.downloadAllLogs(filename = "logs.zip")(wherefilenameis the specified filename for the logs).
Migrating to H2O 3.0
We’re excited about the upcoming release of the latest and greatest version of H2O, and we hope you are too! H2O 3.0 has lots of improvements, including:
- Powerful Python APIs
- Flow, a brand-new intuitive web UI
- The ability to share, annotate, and modify workflows
- Versioned REST APIs with full metadata
- Spark integration using Sparkling Water
- Improved algorithm accuracy and speed
and much more! Overall, H2O has been retooled for better accuracy and performance and to provide additional functionality. If you’re a current user of H2O, we strongly encourage you to upgrade to the latest version to take advantage of the latest features and capabilities.
Please be aware that H2O 3.0 will supersede all previous versions of H2O as the primary version as of May 15th, 2015. Support for previous versions will be offered for a limited time, but there will no longer be any significant updates to the previous version of H2O.
The following information and links will inform you about what’s new and different and help you prepare to upgrade to H2O 3.0.
Overall, H2O 3.0 is more stable, elegant, and simplified, with additional capabilities not available in previous versions of H2O.
Algorithm Changes
Most of the algorithms available in previous versions of H2O have been improved in terms of speed and accuracy. Currently available model types include:
Supervised
- Generalized Linear Model (GLM): Binomial classification, multinomial classification, regression (including logistic regression)
- Distributed Random Forest (DRF): Binomial classification, multinomial classification, regression
- Gradient Boosting Machine (GBM): Binomial classification, multinomial classification, regression
- Deep Learning (DL): Binomial classification, multinomial classification, regression
Unsupervised
- K-means
- Principal Component Analysis
- Autoencoder
There are a few algorithms that are still being refined to provide these same benefits and will be available in a future version of H2O.
Currently, the following algorithms and associated capabilities are still in development:
- Naïve Bayes
Check back for updates, as these algorithms will be re-introduced in an improved form in a future version of H2O.
Note: The SpeeDRF model has been removed, as it was originally intended as an optimization for small data only. This optimization will be added to the Distributed Random Forest model automatically for small data in a future version of H2O.
Parsing Changes
In H2O Classic, the parser reads all the data and tries to guess the column type. In H2O 3.0, the parser reads a subset and makes a type guess for each column. In Flow, you can view the preliminary parse results in the Edit Column Names and Types area. To change the column type, select an option from the drop-down menu to the right of the column. H2O 3.0 can also automatically identify mixed-type columns; in H2O Classic, if one column is mixed integers or real numbers using a string, the output is blank.
Web UI Changes
Our web UI has been completely overhauled with a much more intuitive interface that is similar to IPython Notebook. Each point-and-click action is translated immediately into an individual workflow script that can be saved for later interactive and offline use. As a result, you can now revise and rerun your workflows easily, and can even add comments and rich media.
For more information, refer to our Getting Started with Flow guide, which comprehensively documents how to use Flow. You can also view this brief video, which provides an overview of Flow in action.
API Users
H2O’s new Python API allows Pythonistas to use H2O in their favorite environment. Using the Python command line or an integrated development environment like IPython Notebook, H2O users can control clusters and manage massive datasets quickly.
H2O’s REST API is the basis for the web UI (Flow), as well as the R and Python APIs, and is versioned for stability. It is also easier to understand and use, with full metadata available dynamically from the server, allowing for easier integration by developers.
Java Users
Generated Java REST classes ease REST API use by external programs running in a Java Virtual Machine (JVM).
As in previous versions of H2O, users can export trained models as Java objects for easy integration into JVM applications. H2O is currently the only ML tool that provides this capability, making it the data science tool of choice for enterprise developers.
R Users
If you use H2O primarily in R, be aware that as a result of the improvements to the R package for H2O scripts created using previous versions (Nunes 2.8.6.2 or prior) will require minor revisions to work with H2O 3.0.
To assist our R users in upgrading to H2O 3.0, a “shim” tool has been developed. The shim reviews your script, identifies deprecated or revised parameters and arguments, and suggests replacements.
Note: As of Slater v.3.2.0.10, this shim will no longer be available.
There is also an R Porting Guide that provides a side-by-side comparison of the algorithms in the previous version of H2O with H2O 3.0. It outlines the new, revised, and deprecated parameters for each algorithm, as well as the changes to the output.
Porting R Scripts
This document outlines how to port R scripts written in previous versions of H2O (Nunes 2.8.6.2 or prior, also known as “H2O Classic”) for compatibility with the new H2O 3.0 API. When upgrading from H2O to H2O 3.0, most functions are the same. However, there are some differences that will need to be resolved when porting any scripts that were originally created using H2O to H2O 3.0.
The original R script for H2O is listed first, followed by the updated script for H2O 3.0.
Some of the parameters have been renamed for consistency. For each algorithm, a table that describes the differences is provided.
For additional assistance within R, enter a question mark before the command (for example, ?h2o.glm).
There is also a “shim” available that will review R scripts created with previous versions of H2O, identify deprecated or renamed parameters, and suggest replacements. For more information, refer to the repo here.
Changes from H2O 2.8 to H2O 3.0
- h2o.exec
- h2o.performance
- xval and validation slots
- Principal Components Regression (PCR)
- Saving and Loading Models
h2o.exec
The h2o.exec command is no longer supported. Any workflows using h2o.exec must be revised to remove this command. If the H2O 3.0 workflow contains any parameters or commands from H2O Classic, errors will result and the workflow will fail.
The purpose of h2o.exec was to wrap expressions so that they could be evaluated in a single \Exec2 call. For example,
h2o.exec(fr[,1] + 2/fr[,3])
and
fr[,1] + 2/fr[,3]
produced the same results in H2O. However, the first example makes a single REST call and uses a single temp object, while the second makes several REST calls and uses several temp objects.
Due to the improved architecture in H2O 3.0, the need to use h2o.exec has been eliminated, as the expression can be processed by R as an “unwrapped” typical R expression.
Currently, the only known exception is when factor is used in conjunction with h2o.exec. For example, h2o.exec(fr$myIntCol <- factor(fr$myIntCol)) would become fr$myIntCol <- as.factor(fr$myIntCol)
Note also that an array is not inside a string:
An int array is [1, 2, 3], not “[1, 2, 3]”.
A String array is [“f00”, “b4r”], not “[\”f00\”, \”b4r\”]”
Only string values are enclosed in double quotation marks (").
h2o.performance
To access any exclusively binomial output, use h2o.performance, optionally with the corresponding accessor. The accessor can only use the model metrics object created by h2o.performance. Each accessor is named for its corresponding field (for example, h2o.AUC, h2o.gini, h2o.F1). h2o.performance supports all current algorithms except for K-Means.
If you specify a data frame as a second parameter, H2O will use the specified data frame for scoring. If you do not specify a second parameter, the training metrics for the model metrics object are used.
xval and validation slots
The xval slot has been removed, as nfolds is not currently supported.
The validation slot has been merged with the model slot.
Principal Components Regression (PCR)
Principal Components Regression (PCR) has also been deprecated. To obtain PCR values, create a Principal Components Analysis (PCA) model, then create a GLM model from the scored data from the PCA model.
Saving and Loading Models
Saving and loading a model from R is supported in version 3.0.0.18 and later. H2O 3.0 uses the same binary serialization method as previous versions of H2O, but saves the model and its dependencies into a directory, with each object as a separate file. The save_CV option for available in previous versions of H2O has been deprecated, as h2o.saveAll and h2o.loadAll are not currently supported. The following commands are now supported:
h2o.saveModelh2o.loadModel
Table of Contents
GBM
N-fold cross-validation and grid search are currently supported in H2O 3.0.
Renamed GBM Parameters
The following parameters have been renamed, but retain the same functions:
| H2O Classic Parameter Name | H2O 3.0 Parameter Name |
|---|---|
data |
training_frame |
key |
model_id |
n.trees |
ntrees |
interaction.depth |
max_depth |
n.minobsinnode |
min_rows |
shrinkage |
learn_rate |
n.bins |
nbins |
validation |
validation_frame |
balance.classes |
balance_classes |
max.after.balance.size |
max_after_balance_size |
Deprecated GBM Parameters
The following parameters have been removed:
group_split: Bit-set group splitting of categorical variables is now the default.importance: Variable importances are now computed automatically and displayed in the model output.holdout.fraction: The fraction of the training data to hold out for validation is no longer supported.grid.parallelism: Specifying the number of parallel threads to run during a grid search is no longer supported.
New GBM Parameters
The following parameters have been added:
seed: A random number to control sampling and initialization whenbalance_classesis enabled.score_each_iteration: Display error rate information after each tree in the requested set is built.build_tree_one_node: Run on a single node to use fewer CPUs.
GBM Algorithm Comparison
| H2O Classic | H2O 3.0 |
|---|---|
h2o.gbm <- function( |
h2o.gbm <- function( |
x, |
x, |
y, |
y, |
data, |
training_frame, |
key = "", |
model_id, |
checkpoint |
|
distribution = 'multinomial', |
distribution = c("AUTO", "gaussian", "bernoulli", "multinomial", "poisson", "gamma", "tweedie"), |
tweedie_power = 1.5, |
|
n.trees = 10, |
ntrees = 50 |
interaction.depth = 5, |
max_depth = 5, |
n.minobsinnode = 10, |
min_rows = 10, |
shrinkage = 0.1, |
learn_rate = 0.1, |
sample_rate = 1 |
|
col_sample_rate = 1 |
|
n.bins = 20, |
nbins = 20, |
nbins_top_level, |
|
nbins_cats = 1024, |
|
validation, |
validation_frame = NULL, |
balance.classes = FALSE |
balance_classes = FALSE, |
max.after.balance.size = 5, |
max_after_balance_size = 1, |
seed, |
|
build_tree_one_node = FALSE, |
|
nfolds = 0, |
|
fold_column = NULL, |
|
fold_assignment = c("AUTO", "Random", "Modulo"), |
|
keep_cross_validation_predictions = FALSE, |
|
score_each_iteration = FALSE, |
|
stopping_rounds = 0, |
|
stopping_metric = c("AUTO", "deviance", "logloss", "MSE", "AUC", "r2", "misclassification"), |
|
stopping_tolerance = 0.001, |
|
offset_column = NULL, |
|
weights_column = NULL, |
|
group_split = TRUE, |
|
importance = FALSE, |
|
holdout.fraction = 0, |
|
class.sampling.factors = NULL, |
|
grid.parallelism = 1) |
Output
The following table provides the component name in H2O, the corresponding component name in H2O 3.0 (if supported), and the model type (binomial, multinomial, or all). Many components are now included in h2o.performance; for more information, refer to (h2o.performance).
| H2O Classic | H2O 3.0 | Model Type |
|---|---|---|
@model$priorDistribution |
all |
|
@model$params |
@allparameters |
all |
@model$err |
@model$scoring_history |
all |
@model$classification |
all |
|
@model$varimp |
@model$variable_importances |
all |
@model$confusion |
@model$training_metrics@metrics$cm$table |
binomial and multinomial |
@model$auc |
@model$training_metrics@metrics$AUC |
binomial |
@model$gini |
@model$training_metrics@metrics$Gini |
binomial |
@model$best_cutoff |
binomial |
|
@model$F1 |
@model$training_metrics@metrics$thresholds_and_metric_scores$f1 |
binomial |
@model$F2 |
@model$training_metrics@metrics$thresholds_and_metric_scores$f2 |
binomial |
@model$accuracy |
@model$training_metrics@metrics$thresholds_and_metric_scores$accuracy |
binomial |
@model$error |
binomial |
|
@model$precision |
@model$training_metrics@metrics$thresholds_and_metric_scores$precision |
binomial |
@model$recall |
@model$training_metrics@metrics$thresholds_and_metric_scores$recall |
binomial |
@model$mcc |
@model$training_metrics@metrics$thresholds_and_metric_scores$absolute_MCC |
binomial |
@model$max_per_class_err |
currently replaced by @model$training_metrics@metrics$thresholds_and_metric_scores$min_per_class_correct |
binomial |
GLM
Renamed GLM Parameters
The following parameters have been renamed, but retain the same functions:
| H2O Classic Parameter Name | H2O 3.0 Parameter Name |
|---|---|
data |
training_frame |
key |
model_id |
nlambda |
nlambdas |
lambda.min.ratio |
lambda_min_ratio |
iter.max |
max_iterations |
epsilon |
beta_epsilon |
Deprecated GLM Parameters
The following parameters have been removed:
return_all_lambda: A logical value indicating whether to return every model built during the lambda search. (may be re-added)higher_accuracy: For improved accuracy, adjust thebeta_epsilonvalue.strong_rules: Discards predictors likely to have 0 coefficients prior to model building. (may be re-added as enabled by default)non_negative: Specify a non-negative response. (may be re-added)variable_importances: Variable importances are now computed automatically and displayed in the model output. They have been renamed to Normalized Coefficient Magnitudes.disable_line_search: This parameter has been deprecated, as it was mainly used for testing purposes.max_predictors: Stops training the algorithm if the number of predictors exceeds the specified value. (may be re-added)
New GLM Parameters
The following parameters have been added:
validation_frame: Specify the validation dataset.solver: Select IRLSM or LBFGS.
GLM Algorithm Comparison
| H2O Classic | H2O 3.0 |
|---|---|
h2o.glm <- function( |
h2o.glm( |
x, |
x, |
y, |
y, |
data, |
training_frame, |
key = "", |
model_id, |
validation_frame = NULL |
|
iter.max = 100, |
max_iterations = 50, |
epsilon = 1e-4 |
beta_epsilon = 0 |
strong_rules = TRUE, |
|
return_all_lambda = FALSE, |
|
intercept = TRUE, |
intercept = TRUE |
non_negative = FALSE, |
|
solver = c("IRLSM", "L_BFGS"), |
|
standardize = TRUE, |
standardize = TRUE, |
family, |
family = c("gaussian", "binomial", "multinomial", "poisson", "gamma", "tweedie"), |
link, |
link = c("family_default", "identity", "logit", "log", "inverse", "tweedie"), |
tweedie.p = ifelse(family == "tweedie",1.5, NA_real_) |
tweedie_variance_power = NaN, |
tweedie_link_power = NaN, |
|
alpha = 0.5, |
alpha = 0.5, |
prior = NULL |
prior = 0.0, |
lambda = 1e-5, |
lambda = 1e-05, |
lambda_search = FALSE, |
lambda_search = FALSE, |
nlambda = -1, |
nlambdas = -1, |
lambda.min.ratio = -1, |
lambda_min_ratio = 1.0, |
use_all_factor_levels = FALSE |
use_all_factor_levels = FALSE, |
nfolds = 0, |
nfolds = 0, |
fold_column = NULL, |
|
fold_assignment = c("AUTO", "Random", "Modulo"), |
|
keep_cross_validation_predictions = FALSE, |
|
beta_constraints = NULL, |
beta_constraints = NULL) |
higher_accuracy = FALSE, |
|
variable_importances = FALSE, |
|
disable_line_search = FALSE, |
|
offset = NULL, |
offset_column = NULL, |
weights_column = NULL, |
|
intercept = TRUE, |
|
max_predictors = -1) |
max_active_predictors = -1) |
Output
The following table provides the component name in H2O, the corresponding component name in H2O 3.0 (if supported), and the model type (binomial, multinomial, or all). Many components are now included in h2o.performance; for more information, refer to (h2o.performance).
| H2O Classic | H2O 3.0 | Model Type |
|---|---|---|
@model$params |
@allparameters |
all |
@model$coefficients |
@model$coefficients |
all |
@model$nomalized_coefficients |
@model$coefficients_table$norm_coefficients |
all |
@model$rank |
@model$rank |
all |
@model$iter |
@model$iter |
all |
@model$lambda |
all |
|
@model$deviance |
@model$residual_deviance |
all |
@model$null.deviance |
@model$null_deviance |
all |
@model$df.residual |
@model$residual_degrees_of_freedom |
all |
@model$df.null |
@model$null_degrees_of_freedom |
all |
@model$aic |
@model$AIC |
all |
@model$train.err |
binomial |
|
@model$prior |
binomial |
|
@model$thresholds |
@model$threshold |
binomial |
@model$best_threshold |
binomial |
|
@model$auc |
@model$AUC |
binomial |
@model$confusion |
binomial |
K-Means
Renamed K-Means Parameters
The following parameters have been renamed, but retain the same functions:
| H2O Classic Parameter Name | H2O 3.0 Parameter Name |
|---|---|
data |
training_frame |
key |
model_id |
centers |
k |
cols |
x |
iter.max |
max_iterations |
normalize |
standardize |
Note In H2O, the normalize parameter was disabled by default. The standardize parameter is enabled by default in H2O 3.0 to provide more accurate results for datasets containing columns with large values.
New K-Means Parameters
The following parameters have been added:
userhas been added as an additional option for theinitparameter. Using this parameter forces the K-Means algorithm to start at the user-specified points.user_points: Specify starting points for the K-Means algorithm.
K-Means Algorithm Comparison
| H2O Classic | H2O 3.0 |
|---|---|
h2o.kmeans <- function( |
h2o.kmeans( |
data, |
training_frame, |
cols = '', |
x, |
centers, |
k, |
key = "", |
model_id, |
iter.max = 10, |
max_iterations = 1000, |
normalize = FALSE, |
standardize = TRUE, |
init = "none", |
init = c("Furthest","Random", "PlusPlus"), |
seed = 0, |
seed, |
nfolds = 0, |
|
fold_column = NULL, |
|
fold_assignment = c("AUTO", "Random", "Modulo"), |
|
keep_cross_validation_predictions = FALSE) |
Output
The following table provides the component name in H2O and the corresponding component name in H2O 3.0 (if supported).
| H2O Classic | H2O 3.0 |
|---|---|
@model$params |
@allparameters |
@model$centers |
@model$centers |
@model$tot.withinss |
@model$tot_withinss |
@model$size |
@model$size |
@model$iter |
@model$iterations |
@model$_scoring_history |
|
@model$_model_summary |
Deep Learning
- Renamed Deep Learning Parameters
- Deprecated DL Parameters
- New DL Parameters
- DL Algorithm Comparison
- Output
Note: If the results in the confusion matrix are incorrect, verify that score_training_samples is equal to 0. By default, only the first 10,000 rows are included.
Renamed Deep Learning Parameters
The following parameters have been renamed, but retain the same functions:
| H2O Classic Parameter Name | H2O 3.0 Parameter Name |
|---|---|
data |
training_frame |
key |
model_id |
validation |
validation_frame |
class.sampling.factors |
class_sampling_factors |
override_with_best_model |
overwrite_with_best_model |
dlmodel@model$valid_class_error |
@model$validation_metrics@$MSE |
Deprecated DL Parameters
The following parameters have been removed:
classification: Classification is now inferred from the data type.holdout_fraction: Fraction of the training data to hold out for validation.dlmodel@model$best_cutoff: This output parameter has been removed.
New DL Parameters
The following parameters have been added:
export_weights_and_biases: An additional option allowing users to export the raw weights and biases as H2O frames.
The following options for the loss parameter have been added:
absolute: Provides strong penalties for mispredictionshuber: Can improve results for regression
DL Algorithm Comparison
| H2O Classic | H2O 3.0 |
|---|---|
h2o.deeplearning <- function(x, |
h2o.deeplearning (x, |
y, |
y, |
data, |
training_frame, |
key = "", |
model_id = "", |
override_with_best_model, |
overwrite_with_best_model = true, |
classification = TRUE, |
|
nfolds = 0, |
nfolds = 0 |
validation, |
validation_frame, |
holdout_fraction = 0, |
|
checkpoint = " " |
checkpoint, |
autoencoder, |
autoencoder = false, |
use_all_factor_levels, |
use_all_factor_levels = true |
activation, |
_activation = c("Rectifier", "Tanh", "TanhWithDropout", "RectifierWithDropout", "Maxout", "MaxoutWithDropout"), |
hidden, |
hidden= c(200, 200), |
epochs, |
epochs = 10.0, |
train_samples_per_iteration, |
train_samples_per_iteration = -2, |
target_ratio_comm_to_comp = 0.05 |
|
seed, |
_seed, |
adaptive_rate, |
adaptive_rate = true, |
rho, |
rho = 0.99, |
epsilon, |
epsilon = 1e-08, |
rate, |
rate = .005, |
rate_annealing, |
rate_annealing = 1e-06, |
rate_decay, |
rate_decay = 1.0, |
momentum_start, |
momentum_start = 0, |
momentum_ramp, |
momentum_ramp = 1e+06, |
momentum_stable, |
momentum_stable = 0, |
nesterov_accelerated_gradient, |
nesterov_accelerated_gradient = true, |
input_dropout_ratio, |
input_dropout_ratio = 0.0, |
hidden_dropout_ratios, |
hidden_dropout_ratios, |
l1, |
l1 = 0.0, |
l2, |
l2 = 0.0, |
max_w2, |
max_w2 = Inf, |
initial_weight_distribution, |
initial_weight_distribution = c("UniformAdaptive","Uniform", "Normal"), |
initial_weight_scale, |
initial_weight_scale = 1.0, |
loss, |
loss = "Automatic", "CrossEntropy", "Quadratic", "Absolute", "Huber"), |
distribution = c("AUTO", "gaussian", "bernoulli", "multinomial", "poisson", "gamma", "tweedie", "laplace", "huber"), |
|
tweedie_power = 1.5, |
|
score_interval, |
score_interval = 5, |
score_training_samples, |
score_training_samples = 10000l, |
score_validation_samples, |
score_validation_samples = 0l, |
score_duty_cycle, |
score_duty_cycle = 0.1, |
classification_stop, |
classification_stop = 0 |
regression_stop, |
regression_stop = 1e-6, |
stopping_rounds = 5, |
|
stopping_metric = c("AUTO", "deviance", "logloss", "MSE", "AUC", "r2", "misclassification"), |
|
stopping_tolerance = 0, |
|
quiet_mode, |
quiet_mode = false, |
max_confusion_matrix_size, |
max_confusion_matrix_size, |
max_hit_ratio_k, |
max_hit_ratio_k, |
balance_classes, |
balance_classes = false, |
class_sampling_factors, |
class_sampling_factors, |
max_after_balance_size, |
max_after_balance_size, |
score_validation_sampling, |
score_validation_sampling, |
diagnostics, |
diagnostics = true, |
variable_importances, |
variable_importances = false, |
fast_mode, |
fast_mode = true, |
ignore_const_cols, |
ignore_const_cols = true, |
force_load_balance, |
force_load_balance = true, |
replicate_training_data, |
replicate_training_data = true, |
single_node_mode, |
single_node_mode = false, |
shuffle_training_data, |
shuffle_training_data = false, |
sparse, |
sparse = false, |
col_major, |
col_major = false, |
max_categorical_features, |
max_categorical_features, |
reproducible) |
reproducible=FALSE, |
average_activation |
average_activation = 0, |
sparsity_beta = 0 |
|
export_weights_and_biases=FALSE, |
|
offset_column = NULL, |
|
weights_column = NULL, |
|
nfolds = 0, |
|
fold_column = NULL, |
|
fold_assignment = c("AUTO", "Random", "Modulo"), |
|
keep_cross_validation_predictions = FALSE) |
Output
The following table provides the component name in H2O, the corresponding component name in H2O 3.0 (if supported), and the model type (binomial, multinomial, or all). Many components are now included in h2o.performance; for more information, refer to (h2o.performance).
| H2O Classic | H2O 3.0 | Model Type | |
|---|---|---|---|
@model$priorDistribution |
all |
||
@model$params |
@allparameters |
all |
|
@model$train_class_error |
@model$training_metrics@metrics@$MSE |
all |
|
@model$valid_class_error |
@model$validation_metrics@$MSE |
all |
|
@model$varimp |
@model$_variable_importances |
all |
|
@model$confusion |
@model$training_metrics@metrics$cm$table |
binomial and multinomial |
|
@model$train_auc |
@model$train_AUC |
binomial |
|
@model$_validation_metrics |
all |
||
@model$_model_summary |
all |
||
@model$_scoring_history |
all |
Distributed Random Forest
- Changes to DRF in H2O 3.0
- Renamed DRF Parameters
- Deprecated DRF Parameters
- New DRF Parameters
- DRF Algorithm Comparison
- Output
Changes to DRF in H2O 3.0
Distributed Random Forest (DRF) was represented as h2o.randomForest(type="BigData", ...) in H2O Classic. In H2O Classic, SpeeDRF (type="fast") was not as accurate, especially for complex data with categoricals, and did not address regression problems. DRF (type="BigData") was at least as accurate as SpeeDRF (type="fast") and was the only algorithm that scaled to big data (data too large to fit on a single node).
In H2O 3.0, our plan is to improve the performance of DRF so that the data fits on a single node (optimally, for all cases), which will make SpeeDRF obsolete. Ultimately, the goal is provide a single algorithm that provides the “best of both worlds” for all datasets and use cases.
Please note that H2O does not currently support the ability to specify the number of trees when using h2o.predict for a DRF model.
Note: H2O 3.0 only supports DRF. SpeeDRF is no longer supported. The functionality of DRF in H2O 3.0 is similar to DRF functionality in H2O.
Renamed DRF Parameters
The following parameters have been renamed, but retain the same functions:
| H2O Classic Parameter Name | H2O 3.0 Parameter Name |
|---|---|
data |
training_frame |
key |
model_id |
validation |
validation_frame |
sample.rate |
sample_rate |
ntree |
ntrees |
depth |
max_depth |
balance.classes |
balance_classes |
score.each.iteration |
score_each_iteration |
class.sampling.factors |
class_sampling_factors |
nodesize |
min_rows |
Deprecated DRF Parameters
The following parameters have been removed:
classification: This is now automatically inferred from the response type. To achieve classification with a 0/1 response column, explicitly convert the response to a factor (as.factor()).importance: Variable importances are now computed automatically and displayed in the model output.holdout.fraction: Specifying the fraction of the training data to hold out for validation is no longer supported.doGrpSplit: The bit-set group splitting of categorical variables is now the default.verbose: Infonrmation about tree splits and extra statistics is now included automatically in the stdout.oobee: The out-of-bag error estimate is now computed automatically (if no validation set is specified).stat.type: This parameter was used for SpeeDRF, which is no longer supported.type: This parameter was used for SpeeDRF, which is no longer supported.
New DRF Parameters
The following parameter has been added:
build_tree_one_node: Run on a single node to use fewer CPUs.
DRF Algorithm Comparison
| H2O Classic | H2O 3.0 |
|---|---|
h2o.randomForest <- function(x, |
h2o.randomForest <- function( |
x, |
x, |
y, |
y, |
data, |
training_frame, |
key="", |
model_id, |
validation, |
validation_frame, |
mtries = -1, |
mtries = -1, |
sample.rate=2/3, |
sample_rate = 0.632, |
build_tree_one_node = FALSE, |
|
ntree=50 |
ntrees=50, |
depth=20, |
max_depth = 20, |
min_rows = 1, |
|
nbins=20, |
nbins = 20, |
nbins_top_level, |
|
nbins_cats =1024, |
|
binomial_double_trees = FALSE, |
|
balance.classes = FALSE, |
balance_classes = FALSE, |
seed = -1, |
seed |
nodesize = 1, |
|
classification=TRUE, |
|
importance=FALSE, |
|
weights_column = NULL, |
|
nfolds=0, |
nfolds = 0, |
fold_column = NULL, |
|
fold_assignment = c("AUTO", "Random", "Modulo"), |
|
keep_cross_validation_predictions = FALSE, |
|
score_each_iteration = FALSE, |
|
stopping_rounds = 0, |
|
stopping_metric = c("AUTO", "deviance", "logloss", "MSE", "AUC", "r2", "misclassification"), |
|
stopping_tolerance = 0.001) |
|
holdout.fraction = 0, |
|
max.after.balance.size = 5, |
max_after_balance_size, |
class.sampling.factors = NULL, |
|
doGrpSplit = TRUE, |
|
verbose = FALSE, |
|
oobee = TRUE, |
|
stat.type = "ENTROPY", |
|
type = "fast") |
Output
The following table provides the component name in H2O, the corresponding component name in H2O 3.0 (if supported), and the model type (binomial, multinomial, or all). Many components are now included in h2o.performance; for more information, refer to (h2o.performance).
| H2O Classic | H2O 3.0 | Model Type |
|---|---|---|
@model$priorDistribution |
all |
|
@model$params |
@allparameters |
all |
@model$mse |
@model$scoring_history |
all |
@model$forest |
@model$model_summary |
all |
@model$classification |
all |
|
@model$varimp |
@model$variable_importances |
all |
@model$confusion |
@model$training_metrics@metrics$cm$table |
binomial and multinomial |
@model$auc |
@model$training_metrics@metrics$AUC |
binomial |
@model$gini |
@model$training_metrics@metrics$Gini |
binomial |
@model$best_cutoff |
binomial |
|
@model$F1 |
@model$training_metrics@metrics$thresholds_and_metric_scores$f1 |
binomial |
@model$F2 |
@model$training_metrics@metrics$thresholds_and_metric_scores$f2 |
binomial |
@model$accuracy |
@model$training_metrics@metrics$thresholds_and_metric_scores$accuracy |
binomial |
@model$Error |
@model$Error |
binomial |
@model$precision |
@model$training_metrics@metrics$thresholds_and_metric_scores$precision |
binomial |
@model$recall |
@model$training_metrics@metrics$thresholds_and_metric_scores$recall |
binomial |
@model$mcc |
@model$training_metrics@metrics$thresholds_and_metric_scores$absolute_MCC |
binomial |
@model$max_per_class_err |
currently replaced by @model$training_metrics@metrics$thresholds_and_metric_scores$min_per_class_correct |
binomial |
Github Users
All users who pull directly from the H2O classic repo on Github should be aware that this repo will be renamed. To retain access to the original H2O (2.8.6.2 and prior) repository:
The simple way
This is the easiest way to change your local repo and is recommended for most users.
- Enter
git remote -vto view a list of your repositories. Copy the address your H2O classic repo (refer to the text in brackets below - your address will vary depending on your connection method):
H2O_User-MBP:h2o H2O_User$ git remote -v origin https://{H2O_User@github.com}/h2oai/h2o.git (fetch) origin https://{H2O_User@github.com}/h2oai/h2o.git (push)- Enter
git remote set-url origin {H2O_User@github.com}:h2oai/h2o-2.git, where{H2O_User@github.com}represents the address copied in the previous step.
The more complicated way
This method involves editing the Github config file and should only be attempted by users who are confident enough with their knowledge of Github to do so.
- Enter
vim .git/config. Look for the
[remote "origin"]section:[remote "origin"] url = https://H2O_User@github.com/h2oai/h2o.git fetch = +refs/heads/*:refs/remotes/origin/*- In the
url =line, changeh2o.gittoh2o-2.git. - Save the changes.
The latest version of H2O is stored in the h2o-3 repository. All previous links to this repo will still work, but if you would like to manually update your Github configuration, follow the instructions above, replacing h2o-2 with h2o-3.
POJO Quick Start
This document describes how to build and implement a POJO to use predictive scoring. Java developers should refer to the Javadoc for more information, including packages.
Note: POJOs are not supported for source files larger than 1G. For more information, refer to the FAQ below.
What is a POJO?
H2O allows you to convert the models you have built to a Plain Old Java Object (POJO), which can then be easily deployed within your Java app and scheduled to run on a specified dataset.
POJOs allow users to build a model using H2O and then deploy the model to score in real-time, using the POJO model or a REST API call to a scoring server.
Start H2O in terminal window #1:
$ java -jar h2o.jarBuild a model using your web browser:
- Go to http://localhost:54321
- Click view example Flows near the right edge of the screen. Here is a screenshot of what to look for:
- Click
GBM_Airlines_Classification.flow - If a confirmation prompt appears asking you to “Load Notebook”, click it
- From the “Flow” menu choose the “Run all cells” option
- Scroll down and find the “Model” cell in the notebook. Click on the Download POJO button as shown in the following screenshot:
Note: The instructions below assume that the POJO model was downloaded to the “Downloads” folder.
Download model pieces in a new terminal window - H2O must still be running in terminal window #1:
$ mkdir experiment $ cd experiment $ mv ~/Downloads/gbm_pojo_test.java . $ curl http://localhost:54321/3/h2o-genmodel.jar > h2o-genmodel.jarCreate your main program in terminal window #2 by creating a new file called main.java (
vim main.java) with the following contents:import java.io.*; import hex.genmodel.easy.RowData; import hex.genmodel.easy.EasyPredictModelWrapper; import hex.genmodel.easy.prediction.*; public class main { private static String modelClassName = "gbm_pojo_test"; public static void main(String[] args) throws Exception { hex.genmodel.GenModel rawModel; rawModel = (hex.genmodel.GenModel) Class.forName(modelClassName).newInstance(); EasyPredictModelWrapper model = new EasyPredictModelWrapper(rawModel); RowData row = new RowData(); row.put("Year", "1987"); row.put("Month", "10"); row.put("DayofMonth", "14"); row.put("DayOfWeek", "3"); row.put("CRSDepTime", "730"); row.put("UniqueCarrier", "PS"); row.put("Origin", "SAN"); row.put("Dest", "SFO"); BinomialModelPrediction p = model.predictBinomial(row); System.out.println("Label (aka prediction) is flight departure delayed: " + p.label); System.out.print("Class probabilities: "); for (int i = 0; i < p.classProbabilities.length; i++) { if (i > 0) { System.out.print(","); } System.out.print(p.classProbabilities[i]); } System.out.println(""); } }Compile and run in terminal window 2:
$ javac -cp h2o-genmodel.jar -J-Xmx2g -J-XX:MaxPermSize=128m gbm_pojo_test.java main.java $ java -cp .:h2o-genmodel.jar mainThe following output displays:
Label (aka prediction) is flight departure delayed: YES Class probabilities: 0.4790490513429604,0.5209509486570396
Extracting Models from H2O
Generated models can be extracted from H2O in the following ways:
From the H2O Flow Web UI:
When viewing a model, click the Download POJO button at the top of the model cell, as shown in the example in the Quick start section. You can also preview the POJO inside Flow, but it will only show the first thousand lines or so in the web browser, truncating large models.
From R:
The following code snippet shows an example of H2O building a model and downloading its corresponding POJO from an R script.
library(h2o) h2o.init() path = system.file("extdata", "prostate.csv", package = "h2o") h2o_df = h2o.importFile(path) h2o_df$CAPSULE = as.factor(h2o_df$CAPSULE) model = h2o.glm(y = "CAPSULE", x = c("AGE", "RACE", "PSA", "GLEASON"), training_frame = h2o_df, family = "binomial") h2o.download_pojo(model)
From Python:
The following code snippet shows an example of building a model and downloading its corresponding POJO from a Python script.
import h2o h2o.init() path = h2o.system_file("prostate.csv") h2o_df = h2o.import_file(path) h2o_df['CAPSULE'] = h2o_df['CAPSULE'].asfactor() model = h2o.glm(y = "CAPSULE", x = ["AGE", "RACE", "PSA", "GLEASON"], training_frame = h2o_df, family = "binomial") h2o.download_pojo(model)
Use Cases
The following use cases are demonstrated with code examples:
Reading new data from a CSV file and predicting on it: The PredictCsv class is used by the H2O test harness to make predictions on new data points.
Getting a new observation from a JSON request and returning a prediction
- Calling a user-defined function directly from hive: See the H2O-3 training github repository.
FAQ
How do I score new cases in real-time in a production environment?
If you’re using the UI, click the Preview POJO button for your model. This produces a Java class with methods that you can reference and use in your production app.
What kind of technology would I need to use?
Anything that runs in a JVM. The POJO is a standalone Java class with no dependencies on H2O.
How should I format my data before calling the POJO?
Here are our requirements (assuming you are using the “easy” Prediction API for the POJO as described in the Javadoc).
- Input columns must only contain categorical levels that were seen during training
- Any additional input columns not used for training are ignored
- If no input column is specified, it will be treated as an
NA - Some models do not handle NAs well (e.g., GLM)
- Any transformations applied to data before model training must also be applied before calling the POJO predict method
- Input columns must only contain categorical levels that were seen during training
How do I run a POJO on a Spark Cluster?
The POJO provides just the math logic to do predictions, so you won’t find any Spark (or even H2O) specific code there. If you want to use the POJO to make predictions on a dataset in Spark, create a map to call the POJO for each row and save the result to a new column, row-by-row.
How do I communicate with a remote cluster using the REST API?
You can dl the POJO using the REST API but when calling the POJO predict function, it’s in the same JVM, not across a REST API.
Is it possible to make predictions using my H2O cluster with the REST API?
Yes, but this way of making predictions is separate from the POJO. For more information about in-H2O predictions (as opposed to POJO predictions), see the documentation for the H2O REST API endpoint /3/Predictions.
- Why did I receive the following error when trying to compile the POJO?
Michals-MBP:b michal$ javac -cp h2o-genmodel.jar -J-Xmx2g -J-XX:MaxPermSize=128m drf_b9b9d3be_cf5a_464a_b518_90701549c12a.java
An exception has occurred in the compiler (1.7.0_60). Please file a bug at the Java Developer Connection (http://java.sun.com/webapps/bugreport) after checking the Bug Parade for duplicates. Include your program and the following diagnostic in your report. Thank you.
java.lang.IllegalArgumentException
at java.nio.ByteBuffer.allocate(ByteBuffer.java:330)
at com.sun.tools.javac.util.BaseFileManager$ByteBufferCache.get(BaseFileManager.java:308)
at com.sun.tools.javac.util.BaseFileManager.makeByteBuffer(BaseFileManager.java:280)
at com.sun.tools.javac.file.RegularFileObject.getCharContent(RegularFileObject.java:112)
at com.sun.tools.javac.file.RegularFileObject.getCharContent(RegularFileObject.java:52)
at com.sun.tools.javac.main.JavaCompiler.readSource(JavaCompiler.java:571)
at com.sun.tools.javac.main.JavaCompiler.parse(JavaCompiler.java:632)
at com.sun.tools.javac.main.JavaCompiler.parseFiles(JavaCompiler.java:909)
at com.sun.tools.javac.main.JavaCompiler.compile(JavaCompiler.java:824)
at com.sun.tools.javac.main.Main.compile(Main.java:439)
at com.sun.tools.javac.main.Main.compile(Main.java:353)
at com.sun.tools.javac.main.Main.compile(Main.java:342)
at com.sun.tools.javac.main.Main.compile(Main.java:333)
at com.sun.tools.javac.Main.compile(Main.java:76)
at com.sun.tools.javac.Main.main(Main.java:61)
This error is generated when the source file is larger than 1G.
Grid Search (Hyperparameter Search) API
- REST
- Example
- Grid Search in R
- Grid Search in Python
- Grid Search Java API
- Grid Testing
- Caveats/In Progress
- Documentation
REST
The current implementation of the grid search REST API exposes the following endpoints:
GET /<version>/Grids: List available grids, with optional parameters to sort the list by model metric such as MSEGET /<version>/Grids/<grid_id>: Return specified gridPOST /<version>/Grids/<algo_name>: Start a new grid search<algo_name>: Supported algorithm values are{glm, gbm, drf, kmeans, deeplearning}
Endpoints accept model-specific parameters (e.g., GBMParametersV3 and an additional parameter called hyper_parameters which contains a dictionary of the hyper parameters which will be searched. In this dictionary an array of values is specified for each searched hyperparameter.
{
"ntrees":[1,5],
"learn_rate":[0.1,0.01]
}
An optional search_criteria dictionary specifies options for controlling more advanced search strategies. Currently, full Cartesian is the default. RandomDiscrete allows a random search over the hyperparameter space, with three ways of specifying when to stop the search: max number of models, max time, and metric-based early stopping (e.g., stop if MSE hasn’t improved by 0.0001 over the 5 best models). An example is:
{
"strategy": "RandomDiscrete",
"max_runtime_secs": 600,
"max_models": 100,
"stopping_metric": "AUTO",
"stopping_tolerance": 0.00001,
"stopping_rounds": 5,
"seed": 123456
}
With grid search, each model is built sequentially, allowing users to view each model as it is built.
Example
Invoke a new GBM model grid search by POSTing the following request to /99/Grid/gbm:
parms:{hyper_parameters={"ntrees":[1,5],"learn_rate":[0.1,0.01]}, training_frame="filefd41fe7ac0b_csv_1.hex_2", grid_id="gbm_grid_search", response_column="Species"", ignored_columns=[""]}
Grid Search in R
Grid search in R provides the following capabilities:
H2OGrid class: Represents the results of the grid searchh2o.getGrid(<grid_id>, sort_by, decreasing): Display the specified gridh2o.grid: Start a new grid search parameterized by- model builder name (e.g.,
gbm) - model parameters (e.g.,
ntrees=100) hyper_parametersattribute for passing a list of hyper parameters (e.g.,list(ntrees=c(1,100), learn_rate=c(0.1,0.001)))search_criteriaoptional attribute for specifying more a advanced search strategy
- model builder name (e.g.,
Example
ntrees_opts = c(1, 5)
learn_rate_opts = c(0.1, 0.01)
hyper_parameters = list(ntrees = ntrees_opts, learn_rate = learn_rate_opts)
grid <- h2o.grid("gbm", grid_id="gbm_grid_test", x=1:4, y=5, training_frame=iris.hex, hyper_params = hyper_parameters)
grid_models <- lapply(grid@model_ids, function(mid) {
model = h2o.getModel(mid)
})
Random Hyper-Parameter Grid Search Example
# The following two commands remove any previously installed H2O packages for R.
if ("package:h2o" %in% search()) { detach("package:h2o", unload=TRUE) }
if ("h2o" %in% rownames(installed.packages())) { remove.packages("h2o") }
# Next, we download packages that H2O depends on.
pkgs <- c("methods","statmod","stats","graphics","RCurl","jsonlite","tools","utils")
for (pkg in pkgs) {
if (! (pkg %in% rownames(installed.packages()))) { install.packages(pkg) }
}
# Now we download, install and initialize the H2O package for R.
install.packages("h2o", type="source", repos=(c("http://h2o-release.s3.amazonaws.com/h2o/rel-tukey/7/R")))
library(h2o)
h2o.init(nthreads=-1)
train <- h2o.importFile("http://s3.amazonaws.com/h2o-public-test-data/smalldata/flow_examples/arrhythmia.csv.gz")
dim(train)
response <- 1
predictors <- c(2:ncol(train))
splits<-h2o.splitFrame(train, 0.9, destination_frames = c("trainSplit","validSplit"), seed = 123456)
trainSplit <- splits[[1]]
validSplit <- splits[[2]]
## Hyper-Parameter Search
## Construct a large Cartesian hyper-parameter space
ntrees_opts <- c(10000) ## early stopping will stop earlier
max_depth_opts <- seq(1,20)
min_rows_opts <- c(1,5,10,20,50,100)
learn_rate_opts <- seq(0.001,0.01,0.001)
sample_rate_opts <- seq(0.3,1,0.05)
col_sample_rate_opts <- seq(0.3,1,0.05)
col_sample_rate_per_tree_opts = seq(0.3,1,0.05)
#nbins_cats_opts = seq(100,10000,100) ## no categorical features in this dataset
hyper_params = list( ntrees = ntrees_opts,
max_depth = max_depth_opts,
min_rows = min_rows_opts,
learn_rate = learn_rate_opts,
sample_rate = sample_rate_opts,
col_sample_rate = col_sample_rate_opts,
col_sample_rate_per_tree = col_sample_rate_per_tree_opts
#,nbins_cats = nbins_cats_opts
)
## Search a random subset of these hyper-parmameters (max runtime and max models are enforced, and the search will stop after we don't improve much over the best 5 random models)
search_criteria = list(strategy = "RandomDiscrete", max_runtime_secs = 600, max_models = 100, stopping_metric = "AUTO", stopping_tolerance = 0.00001, stopping_rounds = 5, seed = 123456)
gbm.grid <- h2o.grid("gbm",
grid_id = "mygrid",
x = predictors,
y = response,
# faster to use a 80/20 split
training_frame = trainSplit,
validation_frame = validSplit,
nfolds = 0,
# alternatively, use N-fold cross-validation
#training_frame = train,
#nfolds = 5,
distribution="gaussian", ## best for MSE loss, but can try other distributions ("laplace", "quantile")
## stop as soon as mse doesn't improve by more than 0.1% on the validation set,
## for 2 consecutive scoring events
stopping_rounds = 2,
stopping_tolerance = 1e-3,
stopping_metric = "MSE",
score_tree_interval = 100, ## how often to score (affects early stopping)
seed = 123456, ## seed to control the sampling of the Cartesian hyper-parameter space
hyper_params = hyper_params,
search_criteria = search_criteria)
gbm.sorted.grid <- h2o.getGrid(grid_id = "mygrid", sort_by = "mse")
print(gbm.sorted.grid)
best_model <- h2o.getModel(gbm.sorted.grid@model_ids[[1]])
summary(best_model)
scoring_history <- as.data.frame(best_model@model$scoring_history)
plot(scoring_history$number_of_trees, scoring_history$training_MSE, type="p") #training mse
points(scoring_history$number_of_trees, scoring_history$validation_MSE, type="l") #validation mse
## get the actual number of trees
ntrees <- best_model@model$model_summary$number_of_trees
print(ntrees)
For more information, refer to the R grid search code and runit_GBMGrid_airlines.R.
Grid Search in Python
- Class is
H2OGridSearch <grid_name>.show(): Display a list of models (including model IDs, hyperparameters, and MSE) explored by grid search (where<grid_name>is an instance of anH2OGridSearchclass)grid_search = H2OGridSearch(<model_type), hyper_params=hyper_parameters): Start a new grid search parameterized by:model_typeis the type of H2O estimator model with its unchanged parametershyper_paramsin Python is a dictionary of string parameters (keys) and a list of values to be explored by grid search (values) (e.g.,{'ntrees':[1,100], 'learn_rate':[0.1, 0.001]}search_criteriaoptional dictionary for specifying more a advanced search strategy
Example
hyper_parameters = {'ntrees':[10,50], 'max_depth':[20,10]}
grid_search = H2OGridSearch(H2ORandomForestEstimator, hyper_params=hyper_parameters)
grid_search.train(x=["x1", "x2"], y="y", training_frame=train)
grid_search.show()
For more information, refer to the Python grid search code and pyunit_benign_glm_grid.py.
Grid Search Java API
Each parameter exposed by the schema can specify if it is supported by grid search by specifying the attribute gridable=true in the schema @API annotation. In any case, the Java API does not restrict the parameters supported by grid search.
There are two core entities: Grid and GridSearch. GridSeach is a job-building Grid object and is defined by the user’s model factory and the hyperspace walk strategy. The model factory must be defined for each supported model type (DRF, GBM, DL, and K-means). The hyperspace walk strategy specifies how the user-defined space of hyper parameters is traversed. The space definition is not limited. For each point in hyperspace, model parameters of the specified type are produced.
The implementation supports a simple Cartesian grid search as well as random search with several different stopping criteria. Grid build triggers a new model builder job for each hyperspace point returned by the walk strategy. If the model builder job fails, the resulting model is ignored; however, it can still be tracked in the job list, and errors are returned in the grid build result.
Model builder jobs are run serially in sequential order. More advanced job scheduling schemes are under development. Note that in cases of true big data sequential scheduling will yield the highest performance. It is only with a large cluster and small data that concurrent scheduling will improve performance.
The grid object contains the results of the grid search: a list of model keys produced by the grid search as well as any errors, and a table of metrics for each succesful model. The grid object publishes a simple API to get the models.
Launch the grid search by specifying:
- the common model hyperparameters (parameter values which will be common across all models in the search)
- the search hyperparameters (a map
<parameterName, listOfValues>that defines the parameter spaces to traverse) - optionally, search criteria (an instance of
HyperSpaceSearchCriteria)
The Java API can grid search any parameters defined in the model parameter’s class (e.g., GBMParameters). Paramters that are appropriate for gridding are marked by the @API parameter, but this is not enforced by the framework.
Additional methods are available in the model builder to support creation of model parameters and configuration. This eliminates the requirement of the previous implementation where each gridable value was represented as a double. This also allows users to specify different building strategies for model parameters. For example, the REST layer uses a builder that validates parameters against the model parameter’s schema, where the Java API uses a simple reflective builder. Additional reflections support is provided by PojoUtils (methods setField, getFieldValue).
Example
HashMap<String, Object[]> hyperParms = new HashMap<>();
hyperParms.put("_ntrees", new Integer[]{1, 2});
hyperParms.put("_distribution", new Distribution.Family[]{Distribution.Family.multinomial});
hyperParms.put("_max_depth", new Integer[]{1, 2, 5});
hyperParms.put("_learn_rate", new Float[]{0.01f, 0.1f, 0.3f});
// Setup common model parameters
GBMModel.GBMParameters params = new GBMModel.GBMParameters();
params._train = fr._key;
params._response_column = "cylinders";
// Trigger new grid search job, block for results and get the resulting grid object
GridSearch gs =
GridSearch.startGridSearch(params, hyperParms, GBM_MODEL_FACTORY, new HyperSpaceSearchCriteria.CartesianSearchCriteria());
Grid grid = (Grid) gs.get();
Exposing grid search end-point for a new algorithm
In the following example, the PCA algorithm has been implemented and we would like to expose the algorithm via REST API. The following aspects are assumed:
- The PCA model builder is called
PCA - The PCA parameters are defined in a class called
PCAParameters - The PCA parameters schema is called
PCAParametersV3
To add support for PCA grid search:
Add the PCA model build factory into the
hex.grid.ModelFactoriesclass:class ModelFactories { /* ... */ public static ModelFactory<PCAModel.PCAParameters> PCA_MODEL_FACTORY = new ModelFactory<PCAModel.PCAParameters>() { @Override public String getModelName() { return "PCA"; } @Override public ModelBuilder buildModel(PCAModel.PCAParameters params) { return new PCA(params); } }; }Add the PCA REST end-point schema:
public class PCAGridSearchV99 extends GridSearchSchema<PCAGridSearchHandler.PCAGrid, PCAGridSearchV99, PCAModel.PCAParameters, PCAV3.PCAParametersV3> { }Add the PCA REST end-point handler:
public class PCAGridSearchHandler extends GridSearchHandler<PCAGridSearchHandler.PCAGrid, PCAGridSearchV99, PCAModel.PCAParameters, PCAV3.PCAParametersV3> { public PCAGridSearchV99 train(int version, PCAGridSearchV99 gridSearchSchema) { return super.do_train(version, gridSearchSchema); } @Override protected ModelFactory<PCAModel.PCAParameters> getModelFactory() { return ModelFactories.PCA_MODEL_FACTORY; } @Deprecated public static class PCAGrid extends Grid<PCAModel.PCAParameters> { public PCAGrid() { super(null, null, null, null); } } }Register the REST end-point in the register factory
hex.api.Register:public class Register extends AbstractRegister { @Override public void register() { // ... H2O.registerPOST("/99/Grid/pca", PCAGridSearchHandler.class, "train", "Run grid search for PCA model."); // ... } }
Grid Testing
The current test infrastructure includes:
R Tests
- GBM grids using wine, airlines, and iris datasets verify the consistency of results
- DL grid using the
hiddenparameter verifying the passing of structured parameters as a list of values - Minor R testing support verifying equality of the model’s parameters against a given list of hyper parameters.
JUnit Test
- Basic tests verifying consistency of the results for DRF, GBM, and KMeans
- JUnit test assertions for grid results
There are tests for the RandomDiscrete search criteria in runit_GBMGrid_airlines.R and pyunit_benign_glm_grid.py.
Caveats/In Progress
- Currently, the schema system requires specific classes instead of parameterized classes. For example, the schema definition
Grid<GBMParameters>is not supported unless your define the classGBMGrid extends Grid<GBMParameters>. - Grid Job scheduler is sequential only; schedulers for concurrent builds are under development. Note that in cases of true big data sequential scheduling will yield the highest performance. It is only with a large cluster and small data that concurrent scheduling will improve performance.
- The model builder job and grid jobs are not associated.
- There is no way to list the hyper space parameters that caused a model builder job failure.
Documentation
H2O Core Java Developer Documentation: The definitive Java API guide for the core components of H2O.
H2O Algos Java Developer Documentation: The definitive Java API guide for the algorithms used by H2O.
H2O Architecture
H2O Software Stack
The diagram below shows most of the different components that work together to form the H2O software stack. The diagram is split into a top and bottom section, with the network cloud dividing the two sections.
The top section shows some of the different REST API clients that exist for H2O.
The bottom section shows different components that run within an H2O JVM process.
The color scheme in the diagram shows each layer in a consistent color but always shows user-added customer algorithm code as gray.
REST API Clients
All REST API clients communicate with H2O over a socket connection.
JavaScript
The embedded H2O Web UI is written in JavaScript, and uses the standard
REST API.
R
R scripts can use the H2O R package [‘library(h2o)’]. Users can
write their own R functions than run on H2O with ‘apply’ or ‘ddply’.
Python
Python scripts currently must use the REST API directly. An H2O client
API for python is planned.
Excel
An H2O worksheet for Microsoft Excel is available. It allows you to
import big datasets into H2O and run algorithms like GLM directly from
Excel.
Tableau
Users can pull results from H2O for visualization in Tableau.
Flow H2O Flow is the notebook style Web UI for H2O.
JVM Components
An H2O cloud consists of one or more nodes. Each node is a single JVM process. Each JVM process is split into three layers: language, algorithms, and core infrastructure.
The language layer consists of an expression evaluation engine for R and the Shalala Scala layer. The R evaluation layer is a slave to the R REST client front-end. The Scala layer, however, is a first-class citizen in which you can write native programs and algorithms that use H2O.
The algorithms layer contains those algorithms automatically provided with H2O. These are the parse algorithm used for importing datasets, the math and machine learning algorithms like GLM, and the prediction and scoring engine for model evaluation.
The bottom (core) layer handles resource management. Memory and CPU are managed at this level.
Memory Management
Fluid Vector Frame
A Frame is an H2O Data Frame, the basic unit of data storage exposed
to users. “Fluid Vector” is an internal engineering term that
caught on. It refers to the ability to add and update and remove
columns in a frame “fluidly” (as opposed to the frame being rigid and
immutable). The Frame->Vector->Chunk->Element taxonomy that stores
data in memory is described in Javadoc. The Fluid Vector (or fvec)
code is the column-compressed store implementation.
Distributed K/V store
Atomic and distributed in-memory storage spread across the cluster.
Non-blocking Hash Map
Used in the K/V store implementation.
CPU Management
Job
Jobs are large pieces of work that have progress bars and can be
monitored in the Web UI. Model creation is an example of a job.
MRTask
MRTask stands for MapReduce Task. This is an H2O in-memory MapReduce
Task, not to be confused with a Hadoop MapReduce task.
Fork/Join
A modified JSR166y lightweight task execution framework.
How R (and Python) Interacts with H2O
- How R Scripts Tell H2O to Ingest Data
- How R Scripts Call H2O GLM
- How R Expressions are Sent to H2O for Evaluation
The H2O package for R allows R users to control an H2O cluster from an R script. The R script is a REST API client of the H2O cluster. The data never flows through R.
Note that although these examples are for R, Python and the H2O package for Python behave exactly the same way.
How R Scripts Tell H2O to Ingest Data
- Step 1: The R user calls the importFile function
- Step 2: The R client tells the cluster to read the data
- Step 3: The data is returned from HDFS into a distributed H2O Frame
The following sequence of three steps shows how an R program tells an H2O cluster to read data from HDFS into a distributed H2O Frame.
Step 1: The R user calls the importFile function

Step 2: The R client tells the cluster to read the data
The thin arrows show control information.
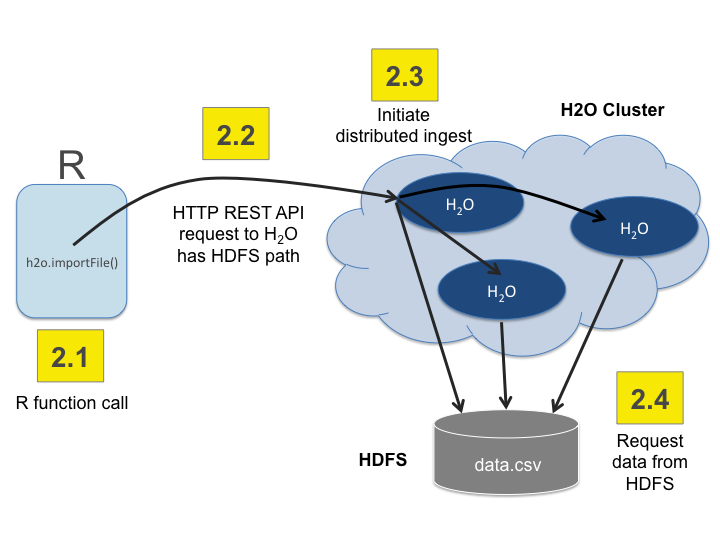
Step 3: The data is returned from HDFS into a distributed H2O Frame
The thin arrows show control information. The thick arrows show data being returned from HDFS. The blocks of data live in the distributed H2O Frame cluster memory.
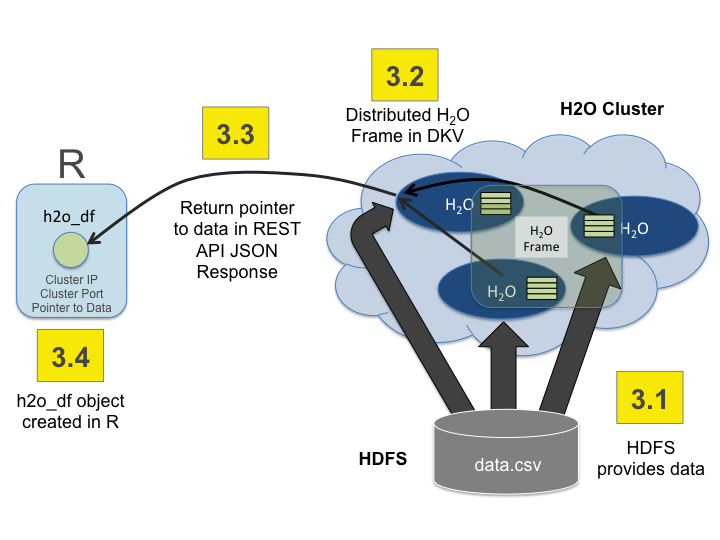
How R Scripts Call H2O GLM
The following diagram shows the different software layers involved when a user runs an R program that starts a GLM on H2O.
The left side shows the steps that run the the R process and the right side shows the steps that run in the H2O cloud. The top layer is the TCP/IP network code that enables the two processes to communicate with each other.
The solid line shows an R->H2O request and the dashed line shows the response for that request.
In the R program, the different components are:
- the R script itself
- the H2O R package
- dependent packages (RCurl, rjson, etc.)
- the R core runtime
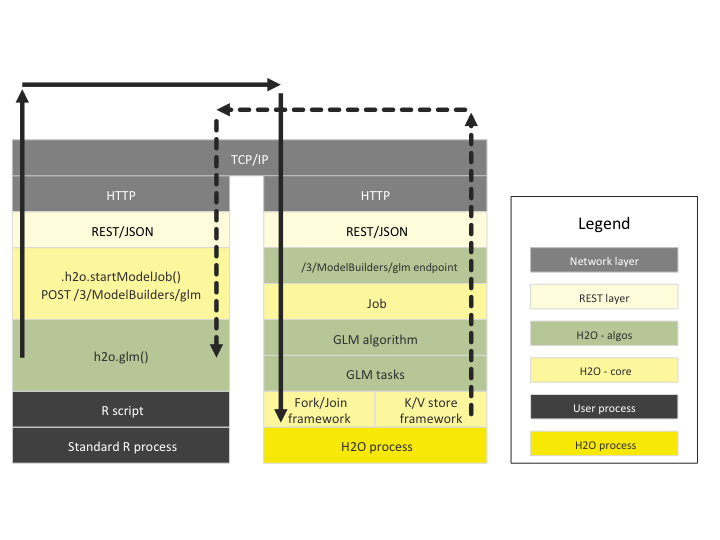
The following diagram shows the R program retrieving the resulting GLM model. (Not shown: the GLM model executing subtasks within H2O and depositing the result into the K/V store or R polling the /3/Jobs URL for the GLM model to complete.)
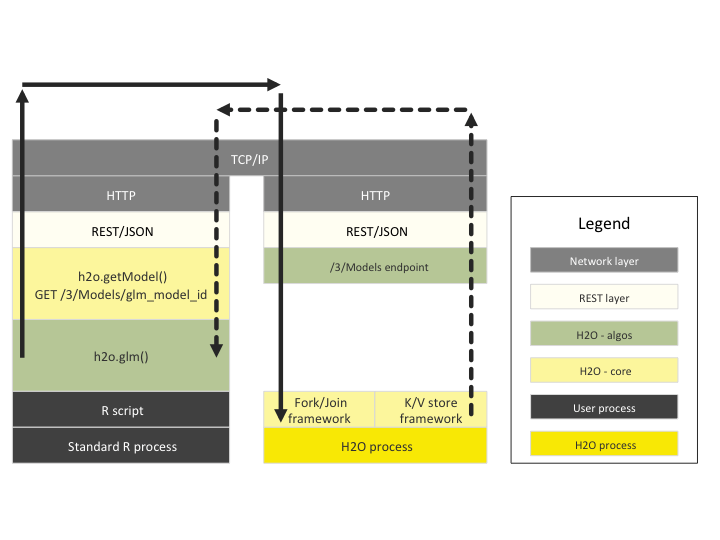
An end-to-end sequence diagram of the same transaction is below. This gives a different perspective of the R and H2O interactions for the same GLM request and the resulting model.
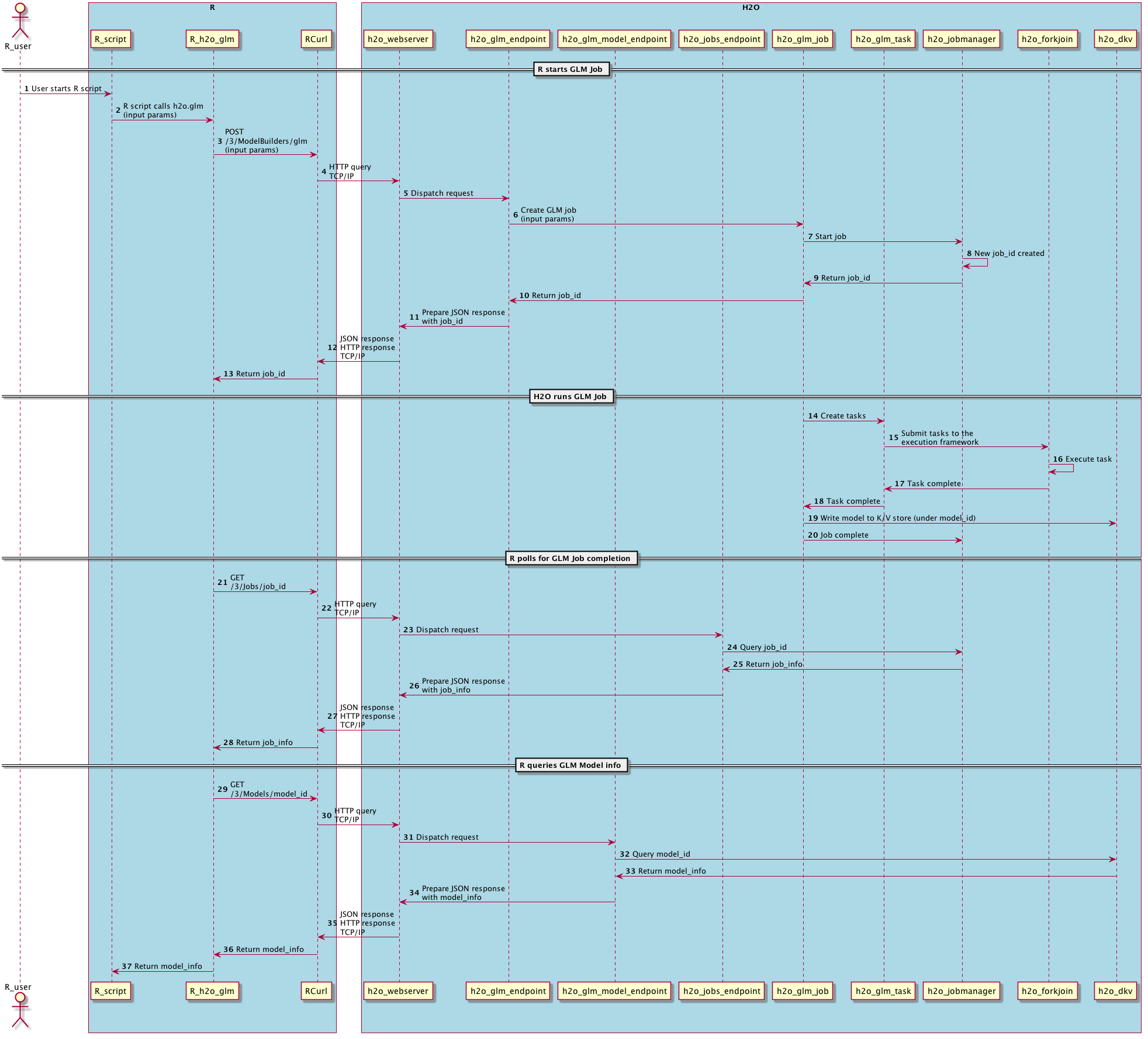
How R Expressions are Sent to H2O for Evaluation
An H2O data frame is represented in R by an S3 object of class
H2OFrame. The S3 object has an id attribute which is a reference to
the big data object inside H2O.
The H2O R package overloads generic operations like ‘summary’ and ‘+’ for this new H2OFrame class. The R core parser makes callbacks into the H2O R package, and these operations get sent to the H2O cluster over an HTTP connection.
The H2O cluster performs the big data operation (for example, ‘+’ on two columns of a dataset imported into H2O) and returns a reference to the result. This reference is stored in a new H2OFrame S3 object inside R.
Complicated expressions are turned into expression trees and evaluated by the Rapids expression engine in the H2O back-end.
Security
H2O contains security features intended for deployment inside a secure data center.
Security model
- Terms
- Assumptions (threat model)
- Data chain-of-custody in a Hadoop data center environment
- What is being secured today
- What is specifically not being secured today
Below is a discussion of what the security assumptions are, and what the H2O software does and does not do.
Terms
| Term | Definition |
|---|---|
| H2O cluster | A collection of H2O nodes that work together. In the H2O Flow Web UI, the cluster status menu item shows the list of nodes in an H2O cluster. |
| H2O node | One JVM instance running the H2O main class. One H2O node corresponds to one OS-level process. In the YARN case, one H2O node corresponds to one mapper instance and one YARN container. |
| H2O embedded web port | Each H2O node contains an embedded web port (by default port 54321). This web port hosts H2O Flow as well as the H2O REST API. The user interacts directly with this web port. |
| H2O internal communication port | Each H2O node also has an internal port (web port+1, so by default port 54322) for internal node-to-node communication. This is a proprietary binary protocol. An attacker using a tool like tcpdump or wireshark may be able to reverse engineer data captured on this communication path. |
Assumptions (threat model)
H2O lives in a secure data center.
Denial of service is not a concern.
- H2O is not designed to withstand a DOS attack.
HTTP traffic between the user client and H2O cluster needs to be encrypted.
- This is true for both interactive sessions (e.g the H2O Flow Web UI) and programmatic sessions (e.g. an R program).
Man-in-the-middle attacks are of low concern.
- Certificate checking on the client side for R/python is not yet implemented.
Internal binary H2O node-to-H2O node traffic does not need to be secured.
- The customer is responsible for the H2O cluster’s perimeter security if this is a concern.
- An example would be putting the nodes for an H2O cluster in a VLAN and opening up one port so user clients can reach the H2O cluster on the embedded web port.
You trust the person that starts H2O to start it correctly.
- Enabling H2O security requires specifying the correct security options.
User client sessions do not need to expire. A session lives at most as long as the cluster lifetime. H2O clusters are started and stopped “frequently enough”.
- All data is stored in-memory, so restarting the H2O cluster wipes all data from memory, and there is nothing to clean from disk.
Once a user is authenticated for access to H2O, they have full access.
- H2O supports authentication but not authorization or access control (ACLs).
H2O clusters are meant to be accessed by only one user.
- Each user starts their own H2O cluster.
- H2O only allows access to the embedded web port to the person that started the cluster.
Data chain-of-custody in a Hadoop data center environment
Notes:
- This holds true for all versions of Hadoop (including YARN) supported by H2O.
Through this sequence, it is shown that a user is only able to access the same data from H2O that they could already access from normal Hadoop jobs.
- Data lives in HDFS
- The files in HDFS have permissions
- An HDFS user has permissions (capabilities) to access certain files
- Kerberos (kinit) can be used to authenticate a user in a Hadoop environment
- A user’s Hadoop MapReduce job inherits the permissions (capabilities) of the user, as well as kinit metadata
- H2O is a Hadoop MapReduce job
- H2O can only access the files in HDFS that the user has permission to access
- Only the user that started the cluster is authenticated for access to the H2O cluster
- The authenticated user can access the same data in H2O that he could access via HDFS
What is being secured today
Standard file permissions security is provided by the Operating System and by HDFS.
The embedded web port in each node of H2O can be secured in two ways:
| Method | Description |
|---|---|
| HTTPS | Encrypted socket communication between the user client and the embedded H2O web port. |
| Authentication | An HTTP Basic Auth username and password from the user client. |
Note: Embedded web port HTTPS and authentication may be used separately or together.
What is specifically not being secured today
- Internal H2O node-to-H2O node communication.
File security in H2O
H2O is a normal user program. Nothing specifically needs to be done by the user to get file security for H2O. Operating System and HDFS permissions “just work”.
Standalone H2O
Since H2O is a regular Java program, the files H2O can access are restricted by the user’s Operating System permissions (capabilities).
H2O on Hadoop
Since H2O is a regular Hadoop MapReduce program, the files H2O can access are restricted by the standard HDFS permissions of the user that starts H2O.
Since H2O is a regular Hadoop MapReduce program, Kerberos (kinit) works seamlessly. (No code was added to H2O to support Kerberos.)
Sparkling Water on YARN
Similar to H2O on Hadoop, this configuration is H2O on Spark on YARN. The YARN job inherits the HDFS permissions of the user.
Embedded web port (by default port 54321) security
For the client side, connection options exist.
For the server side, startup options exist to facilitate security. These are detailed below.
HTTPS
HTTPS client side
Flow Web UI client
When HTTPS is enabled on the server side, the user must provide the https URI scheme to the browser. No http access will exist.
R client
The following code snippet demonstrates connecting to an H2O cluster with HTTPS:
h2o.init(ip = "a.b.c.d", port = 54321, https = TRUE, insecure = TRUE)
The underlying HTTPS implementation is provided by RCurl and by extension libcurl and OpenSSL.
Caution:
Certificate checking has not been implemented yet. The insecure flag tells the client to ignore certificate checking. This means your client is exposed to a man-in-the-middle attack. We assume for the time being that in a secure corporate network such attacks are of low concern. Currently, the insecure flag must be set to TRUE so that in some future version of H2O you will confidently know when certificate checking has actually been implemented.
Python client
Not yet implemented. Please contact H2O for an update.
HTTPS server side
A Java Keystore must be provided on the server side to enable HTTPS. Keystores can be manipulated on the command line with the keytool command.
The underlying HTTPS implementation is provided by Jetty 8 and the Java runtime. (Note: Jetty 8 was chosen to retain Java 6 compatibility.)
Standalone H2O
The following options are available:
-jks <filename>
Java keystore file
-jks_pass <password>
(Default is 'h2oh2o')
Example:
java -jar h2o.jar -jks h2o.jks
H2O on Hadoop
The following options are available:
-jks <filename>
Java keystore file
-jks_pass <password>
(Default is 'h2oh2o')
Example:
hadoop jar h2odriver.jar -n 3 -mapperXmx 10g -jks h2o.jks -output hdfsOutputDirectory
Sparkling Water
The following Spark conf properties exist for Java Keystore configuration:
| Spark conf property | Description |
|---|---|
| spark.ext.h2o.jks | Path to Java Keystore |
| spark.ext.h2o.jks.pass | JKS password |
Example:
$SPARK_HOME/bin/spark-submit --class water.SparklingWaterDriver --conf spark.ext.h2o.jks=/path/to/h2o.jks sparkling-water-assembly-0.2.17-SNAPSHOT-all.jar
Creating your own self-signed Java Keystore
Here is an example of how to create your own self-signed Java Keystore (mykeystore.jks) with a custom keystore password (mypass) and how to run standalone H2O using your Keystore:
# Be paranoid and delete any previously existing keystore.
rm -f mykeystore.jks
# Generate a new keystore.
keytool -genkey -keyalg RSA -keystore mykeystore.jks -storepass mypass -keysize 2048
What is your first and last name?
[Unknown]:
What is the name of your organizational unit?
[Unknown]:
What is the name of your organization?
[Unknown]:
What is the name of your City or Locality?
[Unknown]:
What is the name of your State or Province?
[Unknown]:
What is the two-letter country code for this unit?
[Unknown]:
Is CN=Unknown, OU=Unknown, O=Unknown, L=Unknown, ST=Unknown, C=Unknown correct?
[no]: yes
Enter key password for <mykey>
(RETURN if same as keystore password):
# Run H2O using the newly generated self-signed keystore.
java -jar h2o.jar -jks mykeystore.jks -jks_pass mypass
LDAP authentication
H2O client and server side configuration for LDAP is discussed below. Authentication is implemented using Basic Auth.
LDAP H2O-client side
Flow Web UI client
When authentication is enabled, the user will be presented with a username and password dialog box when attempting to reach Flow.
R client
The following code snippet demonstrates connecting to an H2O cluster with authentication:
h2o.init(ip = "a.b.c.d", port = 54321, username = "myusername", password = "mypassword")
Python client
Not yet implemented. Please contact H2O for an update.
LDAP H2O-server side
An ldap.conf configuration file must be provided by the user. As an example, this file works for H2O’s internal LDAP server. You will certainly need help from your IT security folks to adjust this configuration file for your environment.
Example ldap.conf:
ldaploginmodule {
org.eclipse.jetty.plus.jaas.spi.LdapLoginModule required
debug="true"
useLdaps="false"
contextFactory="com.sun.jndi.ldap.LdapCtxFactory"
hostname="ldap.0xdata.loc"
port="389"
bindDn="cn=admin,dc=0xdata,dc=loc"
bindPassword="0xdata"
authenticationMethod="simple"
forceBindingLogin="true"
userBaseDn="ou=users,dc=0xdata,dc=loc"
userRdnAttribute="uid"
userIdAttribute="uid"
userPasswordAttribute="userPassword"
userObjectClass="inetOrgPerson"
roleBaseDn="ou=groups,dc=0xdata,dc=loc"
roleNameAttribute="cn"
roleMemberAttribute="uniqueMember"
roleObjectClass="groupOfUniqueNames";
};
See the Jetty 8 LdapLoginModule documentation for more information.
Standalone H2O
The following options are available:
-ldap_login
Use Jetty LdapLoginService
-login_conf <filename>
LoginService configuration file
-user_name <username>
Override name of user for which access is allowed
Example:
java -jar h2o.jar -ldap_login -login_conf ldap.conf
java -jar h2o.jar -ldap_login -login_conf ldap.conf -user_name myLDAPusername
H2O on Hadoop
The following options are available:
-ldap_login
Use Jetty LdapLoginService
-login_conf <filename>
LoginService configuration file
-user_name <username>
Override name of user for which access is allowed
Example:
hadoop jar h2odriver.jar -n 3 -mapperXmx 10g -ldap_login -login_conf ldap.conf -output hdfsOutputDirectory
hadoop jar h2odriver.jar -n 3 -mapperXmx 10g -ldap_login -login_conf ldap.conf -user_name myLDAPusername -output hdfsOutputDirectory
Sparkling Water
The following Spark conf properties exist for Java keystore configuration:
| Spark conf property | Description |
|---|---|
| spark.ext.h2o.ldap.login | Use Jetty LdapLoginService |
| spark.ext.h2o.login.conf | LoginService configuration file |
| spark.ext.h2o.user.name | Override name of user for which access is allowed |
Example:
$SPARK_HOME/bin/spark-submit --class water.SparklingWaterDriver --conf spark.ext.h2o.ldap.login=true --conf spark.ext.h2o.login.conf=/path/to/ldap.conf sparkling-water-assembly-0.2.17-SNAPSHOT-all.jar
$SPARK_HOME/bin/spark-submit --class water.SparklingWaterDriver --conf spark.ext.h2o.ldap.login=true --conf spark.ext.h2o.user.name=myLDAPusername --conf spark.ext.h2o.login.conf=/path/to/ldap.conf sparkling-water-assembly-0.2.17-SNAPSHOT-all.jar
Hash file authentication
H2O client and server side configuration for a hardcoded hash file is discussed below. Authentication is implemented using Basic Auth.
Hash file H2O-client side
Flow Web UI client
When authentication is enabled, the user will be presented with a username and password dialog box when attempting to reach Flow.
R client
The following code snippet demonstrates connecting to an H2O cluster with authentication:
h2o.init(ip = "a.b.c.d", port = 54321, username = "myusername", password = "mypassword")
Python client
Not yet implemented. Please contact H2O for an update.
Hash file H2O-server side
A realm.properties configuration file must be provided by the user.
Example realm.properties:
# See https://wiki.eclipse.org/Jetty/Howto/Secure_Passwords
# java -cp h2o.jar org.eclipse.jetty.util.security.Password
username1: password1
username2: MD5:6cb75f652a9b52798eb6cf2201057c73
Generate secure passwords using the Jetty secure password generation tool:
java -cp h2o.jar org.eclipse.jetty.util.security.Password username password
See the Jetty 8 HashLoginService documentation and Jetty 8 Secure Password HOWTO for more information.
Standalone H2O
The following options are available:
-hash_login
Use Jetty HashLoginService
-login_conf <filename>
LoginService configuration file
Example:
java -jar h2o.jar -hash_login -login_conf realm.properties
H2O on Hadoop
The following options are available:
-hash_login
Use Jetty HashLoginService
-login_conf <filename>
LoginService configuration file
Example:
hadoop jar h2odriver.jar -n 3 -mapperXmx 10g -hash_login -login_conf realm.propertes -output hdfsOutputDirectory
Sparkling Water
The following Spark conf properties exist for hash login service configuration:
| Spark conf property | Description |
|---|---|
| spark.ext.h2o.hash.login | Use Jetty HashLoginService |
| spark.ext.h2o.login.conf | LoginService configuration file |
Example:
$SPARK_HOME/bin/spark-submit --class water.SparklingWaterDriver --conf spark.ext.h2o.hash.login=true --conf spark.ext.h2o.login.conf=/path/to/realm.properties sparkling-water-assembly-0.2.17-SNAPSHOT-all.jar
FAQ
- General Troubleshooting Tips
- Algorithms
- Building H2O
- Clusters
- Data
- General
- Hadoop
- Java
- Python
- R
- Sparkling Water
- Tunneling between servers with H2O
General Troubleshooting Tips
Confirm your internet connection is active.
Test connectivity using curl: First, log in to the first node and enter
curl http://<Node2IP>:54321(where<Node2IP>is the IP address of the second node. Then, log in to the second node and entercurl http://<Node1IP>:54321(where<Node1IP>is the IP address of the first node). Look for output from H2O.Try allocating more memory to H2O by modifying the
-Xmxvalue when launching H2O from the command line (for example,java -Xmx10g -jar h2o.jarallocates 10g of memory for H2O). If you create a cluster with four 20g nodes (by specifying-Xmx20gfour times), H2O will have a total of 80 gigs of memory available. For best performance, we recommend sizing your cluster to be about four times the size of your data. To avoid swapping, the-Xmxallocation must not exceed the physical memory on any node. Allocating the same amount of memory for all nodes is strongly recommended, as H2O works best with symmetric nodes.Confirm that no other sessions of H2O are running. To stop all running H2O sessions, enter
ps -efww | grep h2oin your shell (OSX or Linux).- Confirm ports 54321 and 54322 are available for both TCP and UDP. Launch Telnet (for Windows users) or Terminal (for OS X users), then type
telnet localhost 54321,telnet localhost 54322 - Confirm your firewall is not preventing the nodes from locating each other. If you can’t launch H2O, we recommend temporarily disabling any firewalls until you can confirm they are not preventing H2O from launching.
- Confirm the nodes are not using different versions of H2O. If the H2O initialization is not successful, look at the output in the shell - if you see
Attempting to join /localhost:54321 with an H2O version mismatch (md5 differs), update H2O on all the nodes to the same version. - Confirm that there is space in the
/tmpdirectory.- Windows: In Command Prompt, enter
TEMPand%TEMP%and delete files as needed, or use Disk Cleanup. - OS X: In Terminal, enter
open $TMPDIRand delete the folder with your username.
- Windows: In Command Prompt, enter
- Confirm that the username is the same on all nodes; if not, define the cloud in the terminal when launching using
-name:java -jar h2o.jar -name myCloud. - Confirm that there are no spaces in the file path name used to launch H2O.
- Confirm that the nodes are not on different networks by confirming that the IP addresses of the nodes are the same in the output:
INFO: Listening for HTTP and REST traffic on IP_Address/ 06-18 10:54:21.586 192.168.1.70:54323 25638 main INFO: H2O cloud name: 'H2O_User' on IP_Address, discovery address /Discovery_Address INFO: Cloud of size 1 formed [IP_Address] - Check if the nodes have different interfaces; if so, use the -network option to define the network (for example,
-network 127.0.0.1). To use a network range, use a comma to separate the IP addresses (for example,-network 123.45.67.0/22,123.45.68.0/24). - Force the bind address using
-ip:java -jar h2o.jar -ip <IP_Address> -port <PortNumber>. - (Hadoop only) Try launching H2O with a longer timeout:
hadoop jar h2odriver.jar -timeout 1800 - (Hadoop only) Try to launch H2O using more memory:
hadoop jar h2odriver.jar -mapperXmx 10g. The cluster’s memory capacity is the sum of all H2O nodes in the cluster. - (Linux only) Check if you have SELINUX or IPTABLES enabled; if so, disable them.
- (EC2 only) Check the configuration for the EC2 security group.
The following error message displayed when I tried to launch H2O - what should I do?
Exception in thread "main" java.lang.UnsupportedClassVersionError: water/H2OApp
: Unsupported major.minor version 51.0
at java.lang.ClassLoader.defineClass1(Native Method)
at java.lang.ClassLoader.defineClassCond(Unknown Source)
at java.lang.ClassLoader.defineClass(Unknown Source)
at java.security.SecureClassLoader.defineClass(Unknown Source)
at java.net.URLClassLoader.defineClass(Unknown Source)
at java.net.URLClassLoader.access$000(Unknown Source)
at java.net.URLClassLoader$1.run(Unknown Source)
at java.security.AccessController.doPrivileged(Native Method)
at java.net.URLClassLoader.findClass(Unknown Source)
at java.lang.ClassLoader.loadClass(Unknown Source)
at sun.misc.Launcher$AppClassLoader.loadClass(Unknown Source)
at java.lang.ClassLoader.loadClass(Unknown Source)
Could not find the main class: water.H2OApp. Program will exit.
This error output indicates that your Java version is not supported. Upgrade to Java 7 (JVM) or later and H2O should launch successfully.
Algorithms
What does it mean if the r2 value in my model is negative?
The coefficient of determination (also known as r^2) can be negative if:
- linear regression is used without an intercept (constant)
- non-linear functions are fitted to the data
- predictions compared to the corresponding outcomes are not based on the model-fitting procedure using those data
- it is early in the build process (may self-correct as more trees are added)
If your r2 value is negative after your model is complete, your model is likely incorrect. Make sure your data is suitable for the type of model, then try adding an intercept.
What’s the process for implementing new algorithms in H2O?
This blog post by Cliff walks you through building a new algorithm, using K-Means, Quantiles, and Grep as examples.
To learn more about performance characteristics when implementing new algorithms, refer to Cliff’s KV Store Guide.
How do I find the standard errors of the parameter estimates (p-values)?
P-values are currently supported for non-regularized GLM. The following requirements must be met:
- The family cannot be multinomial
- The lambda value must be equal to zero
- The IRLSM solver must be used
- Lambda search cannot be used
To generate p-values, do one of the following:
- check the compute_p_values checkbox in the GLM model builder in Flow
- use
compute_p_values=TRUEin R or Python while creating the model
The p-values are listed in the coefficients table (as shown in the following example screenshot):
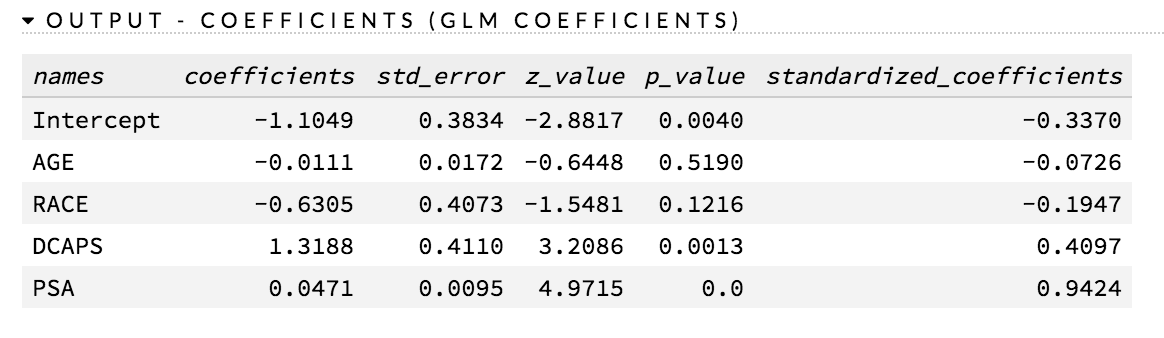
How do I specify regression or classification for Distributed Random Forest in the web UI?
If the response column is numeric, H2O generates a regression model. If the response column is enum, the model uses classification. To specify the column type, select it from the drop-down column name list in the Edit Column Names and Types section during parsing.
What’s the largest number of classes that H2O supports for multinomial prediction?
For tree-based algorithms, the maximum number of classes (or levels) for a response column is 1000.
How do I obtain a tree diagram of my DRF model?
Output the SVG code for the edges and nodes. A simple tree visitor is available here and the Java code generator is available here.
Is Word2Vec available? I can see the Java and R sources, but calling the API generates an error.
Word2Vec, along with other natural language processing (NLP) algos, are currently in development in the current version of H2O.
What are the “best practices” for preparing data for a K-Means model?
There aren’t specific “best practices,” as it depends on your data and the column types. However, removing outliers and transforming any categorical columns to have the same weight as the numeric columns will help, especially if you’re standardizing your data.
What is your implementation of Deep Learning based on?
Our Deep Learning algorithm is based on the feedforward neural net. For more information, refer to our Data Science documentation or Wikipedia.
How is deviance computed for a Deep Learning regression model?
For a Deep Learning regression model, deviance is computed as follows:
Loss = MeanSquare -> MSE==Deviance For Absolute/Laplace or Huber -> MSE != Deviance.
For my 0-tree GBM multinomial model, I got a different score depending on whether or not validation was enabled, even though my dataset was the same - why is that?
Different results may be generated because of the way H2O computes the initial MSE.
How does your Deep Learning Autoencoder work? Is it deep or shallow?
H2O’s DL autoencoder is based on the standard deep (multi-layer) neural net architecture, where the entire network is learned together, instead of being stacked layer-by-layer. The only difference is that no response is required in the input and that the output layer has as many neurons as the input layer. If you don’t achieve convergence, then try using the Tanh activation and fewer layers. We have some example test scripts here, and even some that show how stacked auto-encoders can be implemented in R.
Are there any H2O examples using text for classification?
Currently, the following examples are available for Sparkling Water:
a) Use TF-IDF weighting scheme for classifying text messages https://github.com/h2oai/sparkling-water/blob/master/examples/scripts/mlconf_2015_hamSpam.script.scala
b) Use Word2Vec Skip-gram model + GBM for classifying job titles https://github.com/h2oai/sparkling-water/blob/master/examples/scripts/craigslistJobTitles.scala
Most machine learning tools cannot predict with a new categorical level that was not included in the training set. How does H2O make predictions in this scenario?
Here is an example of how the prediction process works in H2O:
- Train a model using data that has a categorical predictor column with levels B,C, and D (no other levels); this level will be the “training set domain”: {B,C,D}
- During scoring, the test set has only rows with levels A,C, and E for that column; this is the “test set domain”: {A,C,E}
- For scoring, a combined “scoring domain” is created, which is the training domain appended with the extra test set domain entries: {B,C,D,A,E}
- Each model can handle these extra levels {A,E} separately during scoring.
The behavior for unseen categorical levels depends on the algorithm and how it handles missing levels (NA values):
- DRF and GBM treat missing or NA factor levels as the smallest value present (left-most in the bins), which can go left or right for any split. Unseen factor levels always go left in any split.
- Deep Learning creates an extra input neuron for missing and unseen categorical levels, which can remain untrained if there were no missing or unseen categorical levels in the training data, resulting in a random contribution to the next layer during testing.
- GLM skips unseen levels in the beta*x dot product.
How are quantiles computed?
The quantile results in Flow are computed lazily on-demand and cached. It is a fast approximation (max - min / 1024) that is very accurate for most use cases.
If the distribution is skewed, the quantile results may not be as accurate as the results obtained using h2o.quantile in R or H2OFrame.quantile in Python.
How do I create a classification model? The model always defaults to regression.
To create a classification model, the response column type must be enum - if the response is numeric, a regression model is created.
To convert the response column:
Before parsing, click the drop-down menu to the right of the column name or number and select
Enum
or
Click on the .hex link for the data frame (or use the
getFrameSummary "<frame_name>.hex"command, where<frame_name>is the name of the frame), then click the Convert to enum link to the right of the column name or number
Building H2O
During the build process, the following error message displays. What do I need to do to resolve it?
Error: Missing name at classes.R:19
In addition: Warning messages:
1: @S3method is deprecated. Please use @export instead
2: @S3method is deprecated. Please use @export instead
Execution halted
To build H2O, Roxygen2 version 4.1.1 is required.
To update your Roxygen2 version, install the versions package in R, then use install.versions("roxygen2", "4.1.1").
Using ./gradlew build doesn’t generate a build successfully - is there anything I can do to troubleshoot?
Use ./gradlew clean before running ./gradlew build.
I tried using ./gradlew build after using git pull to update my local H2O repo, but now I can’t get H2O to build successfully - what should I do?
Try using ./gradlew build -x test - the build may be failing tests if data is not synced correctly.
Clusters
When trying to launch H2O, I received the following error message: ERROR: Too many retries starting cloud. What should I do?
If you are trying to start a multi-node cluster where the nodes use multiple network interfaces, by default H2O will resort to using the default host (127.0.0.1).
To specify an IP address, launch H2O using the following command:
java -jar h2o.jar -ip <IP_Address> -port <PortNumber>
If this does not resolve the issue, try the following additional troubleshooting tips:
Confirm your internet connection is active.
Test connectivity using curl: First, log in to the first node and enter curl http://
:54321 (where is the IP address of the second node. Then, log in to the second node and enter curl http:// :54321 (where is the IP address of the first node). Look for output from H2O. Confirm ports 54321 and 54322 are available for both TCP and UDP.
- Confirm your firewall is not preventing the nodes from locating each other.
- Confirm the nodes are not using different versions of H2O.
- Confirm that the username is the same on all nodes; if not, define the cloud in the terminal when launching using
-name:java -jar h2o.jar -name myCloud. - Confirm that the nodes are not on different networks.
- Check if the nodes have different interfaces; if so, use the -network option to define the network (for example,
-network 127.0.0.1). - Force the bind address using
-ip:java -jar h2o.jar -ip <IP_Address> -port <PortNumber>. - (Linux only) Check if you have SELINUX or IPTABLES enabled; if so, disable them.
- (EC2 only) Check the configuration for the EC2 security group.
What should I do if I tried to start a cluster but the nodes started independent clouds that are not connected?
Because the default cloud name is the user name of the node, if the nodes are on different operating systems (for example, one node is using Windows and the other uses OS X), the different user names on each machine will prevent the nodes from recognizing that they belong to the same cloud. To resolve this issue, use -name to configure the same name for all nodes.
One of the nodes in my cluster is unavailable — what do I do?
H2O does not support high availability (HA). If a node in the cluster is unavailable, bring the cluster down and create a new healthy cluster.
How do I add new nodes to an existing cluster?
New nodes can only be added if H2O has not started any jobs. Once H2O starts a task, it locks the cluster to prevent new nodes from joining. If H2O has started a job, you must create a new cluster to include additional nodes.
How do I check if all the nodes in the cluster are healthy and communicating?
In the Flow web UI, click the Admin menu and select Cluster Status.
How do I create a cluster behind a firewall?
H2O uses two ports:
- The
REST_APIport (54321): Specify when launching H2O using-port; uses TCP only. - The
INTERNAL_COMMUNICATIONport (54322): Implied based on the port specified as theREST_APIport, +1; requires TCP and UDP.
You can start the cluster behind the firewall, but to reach it, you must make a tunnel to reach the REST_API port. To use the cluster, the REST_API port of at least one node must be reachable.
I launched H2O instances on my nodes - why won’t they form a cloud?
If you launch without specifying the IP address by adding argument -ip:
$ java -Xmx20g -jar h2o.jar -flatfile flatfile.txt -port 54321
and multiple local IP addresses are detected, H2O uses the default localhost (127.0.0.1) as shown below:
10:26:32.266 main WARN WATER: Multiple local IPs detected:
+ /198.168.1.161 /198.168.58.102
+ Attempting to determine correct address...
10:26:32.284 main WARN WATER: Failed to determine IP, falling back to localhost.
10:26:32.325 main INFO WATER: Internal communication uses port: 54322
+ Listening for HTTP and REST traffic
+ on http://127.0.0.1:54321/
10:26:32.378 main WARN WATER: Flatfile configuration does not include self:
/127.0.0.1:54321 but contains [/192.168.1.161:54321, /192.168.1.162:54321]
To avoid using 127.0.0.1 on servers with multiple local IP addresses, run the command with the -ip argument to force H2O to launch at the specified IP:
$ java -Xmx20g -jar h2o.jar -flatfile flatfile.txt -ip 192.168.1.161 -port 54321
How does the timeline tool work?
The timeline is a debugging tool that provides information on the current communication between H2O nodes. It shows a snapshot of the most recent messages passed between the nodes. Each node retains its own history of messages sent to or received from other nodes.
H2O collects these messages from all the nodes and orders them by whether they were sent or received. Each node has an implicit internal order where sent messages must precede received messages on the other node.
The following information displays for each message:
HH:MM:SS:MSandnanosec: The local time of the eventWho: The endpoint of the message; can be either a source/receiver node or source node and multicast for broadcasted messagesI/O Type: The type of communication (either UDP for small messages or TCP for large messages)Note: UDP messages are only sent if the UDP option was enabled when launching H2O or for multicast when a flatfile is not used for configuration.
Event: The type of H2O message. The most common type is a distributed task, which displays asexec(the requested task) ->ack(results of the processed task) ->ackck(sender acknowledges receiving the response, task is completed and removed)rebooted: Sent during node startupheartbeat: Provides small message tracking information about node health, exchanged periodically between nodesfetchack: Aknowledgement of theFetchtype task, which retrieves the ID of a previously unseen typebytes: Information extracted from the message, including the type of the task and the unique task number
Data
How should I format my SVMLight data before importing?
The data must be formatted as a sorted list of unique integers, the column indices must be >= 1, and the columns must be in ascending order.
What date and time formats does H2O support?
H2O is set to auto-detect two major date/time formats. Because many date time formats are ambiguous (e.g. 01/02/03), general date time detection is not used.
The first format is for dates formatted as yyyy-MM-dd. Year is a four-digit number, the month is a two-digit number ranging from 1 to 12, and the day is a two-digit value ranging from 1 to 31. This format can also be followed by a space and then a time (specified below).
The second date format is for dates formatted as dd-MMM-yy. Here the day must be one or two digits with a value ranging from 1 to 31. The month must be either a three-letter abbreviation or the full month name but is not case sensitive. The year must be either two or four digits. In agreement with POSIX standards, two-digit dates >= 69 are assumed to be in the 20th century (e.g. 1969) and the rest are part of the 21st century. This date format can be followed by either a space or colon character and then a time. The ‘-‘ between the values is optional.
Times are specified as HH:mm:ss. HH is a two-digit hour and must be a value between 0-23 (for 24-hour time) or 1-12 (for a twelve-hour clock). mm is a two-digit minute value and must be a value between 0-59. ss is a two-digit second value and must be a value between 0-59. This format can be followed with either milliseconds, nanoseconds, and/or the cycle (i.e. AM/PM). If milliseconds are included, the format is HH:mm:ss:SSS. If nanoseconds are included, the format is HH:mm:ss:SSSnnnnnn. H2O only stores fractions of a second up to the millisecond, so accuracy may be lost. Nanosecond parsing is only included for convenience. Finally, a valid time can end with a space character and then either “AM” or “PM”. For this format, the hours must range from 1 to 12. Within the time, the ‘:’ character can be replaced with a ‘.’ character.
How does H2O handle name collisions/conflicts in the dataset?
If there is a name conflict (for example, column 48 isn’t named, but C48 already exists), then the column name in concatenated to itself until a unique name is created. So for the previously cited example, H2O will try renaming the column to C48C48, then C48C48C48, and so on until an unused name is generated.
What types of data columns does H2O support?
Currently, H2O supports:
- float (any IEEE double)
- integer (up to 64bit, but compressed according to actual range)
- factor (same as integer, but with a String mapping, often handled differently in the algorithms)
- time (same as 64bit integer, but with a time-since-Unix-epoch interpretation)
- UUID (128bit integer, no math allowed)
- String
I am trying to parse a Gzip data file containing multiple files, but it does not parse as quickly as the uncompressed files. Why is this?
Parsing Gzip files is not done in parallel, so it is sequential and uses only one core. Other parallel parse compression schemes are on the roadmap.
General
How do I score using an exported JSON model?
Since JSON is just a representation format, it cannot be directly executed, so a JSON export can’t be used for scoring. However, you can score by:
including the POJO in your execution stream and handing it observations one at a time
or
handing your data in bulk to an H2O cluster, which will score using high throughput parallel and distributed bulk scoring.
How do I score using an exported POJO?
The generated POJO can be used indepedently of a H2O cluster. First use curl to send the h2o-genmodel.jar file and the java code for model to the server. The following is an example; the ip address and model names will need to be changed.
mkdir tmpdir
cd tmpdir
curl http://127.0.0.1:54321/3/h2o-genmodel.jar > h2o-genmodel.jar
curl http://127.0.0.1:54321/3/Models.java/gbm_model > gbm_model.java
To score a simple .CSV file, download the PredictCSV.java file and compile it with the POJO. Make a subdirectory for the compilation (this is useful if you have multiple models to score on).
wget https://raw.githubusercontent.com/h2oai/h2o-3/master/h2o-r/tests/testdir_javapredict/PredictCSV.java
mkdir gbm_model_dir
javac -cp h2o-genmodel.jar -J-Xmx2g -J-XX:MaxPermSize=128m PredictCSV.java gbm_model.java -d gbm_model_dir
Specify the following:
- the classpath using
-cp - the model name (or class) using
--model - the csv file you want to score using
--input - the location for the predictions using
--output.
You must match the table column names to the order specified in the POJO. The output file will be in a .hex format, which is a lossless text representation of floating point numbers. Both R and Java will be able to read the hex strings as numerics.
java -ea -cp h2o-genmodel.jar:gbm_model_dir -Xmx4g -XX:MaxPermSize=256m -XX:ReservedCodeCacheSize=256m PredictCSV --header --model gbm_model --input input.csv --output output.csv
How do I predict using multiple response variables?
Currently, H2O does not support multiple response variables. To predict different response variables, build multiple models.
How do I kill any running instances of H2O?
In Terminal, enter ps -efww | grep h2o, then kill any running PIDs. You can also find the running instance in Terminal and press Ctrl + C on your keyboard. To confirm no H2O sessions are still running, go to http://localhost:54321 and verify that the H2O web UI does not display.
Why is H2O not launching from the command line?
$ java -jar h2o.jar &
% Exception in thread "main" java.lang.ExceptionInInitializerError
at java.lang.Class.initializeClass(libgcj.so.10)
at water.Boot.getMD5(Boot.java:73)
at water.Boot.<init>(Boot.java:114)
at water.Boot.<clinit>(Boot.java:57)
at java.lang.Class.initializeClass(libgcj.so.10)
Caused by: java.lang.IllegalArgumentException
at java.util.regex.Pattern.compile(libgcj.so.10)
at water.util.Utils.<clinit>(Utils.java:1286)
at java.lang.Class.initializeClass(libgcj.so.10)
...4 more
The only prerequisite for running H2O is a compatible version of Java. We recommend Oracle’s Java 1.7.
Why did I receive the following error when I tried to launch H2O?
[root@sandbox h2o-dev-0.3.0.1188-hdp2.2]hadoop jar h2odriver.jar -nodes 2 -mapperXmx 1g -output hdfsOutputDirName
Determining driver host interface for mapper->driver callback...
[Possible callback IP address: 10.0.2.15]
[Possible callback IP address: 127.0.0.1]
Using mapper->driver callback IP address and port: 10.0.2.15:41188
(You can override these with -driverif and -driverport.)
Memory Settings:
mapreduce.map.java.opts: -Xms1g -Xmx1g -Dlog4j.defaultInitOverride=true
Extra memory percent: 10
mapreduce.map.memory.mb: 1126
15/05/08 02:33:40 INFO impl.TimelineClientImpl: Timeline service address: http://sandbox.hortonworks.com:8188/ws/v1/timeline/
15/05/08 02:33:41 INFO client.RMProxy: Connecting to ResourceManager at sandbox.hortonworks.com/10.0.2.15:8050
15/05/08 02:33:47 INFO mapreduce.JobSubmitter: number of splits:2
15/05/08 02:33:48 INFO mapreduce.JobSubmitter: Submitting tokens for job: job_1431052132967_0001
15/05/08 02:33:51 INFO impl.YarnClientImpl: Submitted application application_1431052132967_0001
15/05/08 02:33:51 INFO mapreduce.Job: The url to track the job: http://sandbox.hortonworks.com:8088/proxy/application_1431052132967_0001/
Job name 'H2O_3889' submitted
JobTracker job ID is 'job_1431052132967_0001'
For YARN users, logs command is 'yarn logs -applicationId application_1431052132967_0001'
Waiting for H2O cluster to come up...
H2O node 10.0.2.15:54321 requested flatfile
ERROR: Timed out waiting for H2O cluster to come up (120 seconds)
ERROR: (Try specifying the -timeout option to increase the waiting time limit)
15/05/08 02:35:59 INFO impl.TimelineClientImpl: Timeline service address: http://sandbox.hortonworks.com:8188/ws/v1/timeline/
15/05/08 02:35:59 INFO client.RMProxy: Connecting to ResourceManager at sandbox.hortonworks.com/10.0.2.15:8050
----- YARN cluster metrics -----
Number of YARN worker nodes: 1
----- Nodes -----
Node: http://sandbox.hortonworks.com:8042 Rack: /default-rack, RUNNING, 1 containers used, 0.2 / 2.2 GB used, 1 / 8 vcores used
----- Queues -----
Queue name: default
Queue state: RUNNING
Current capacity: 0.11
Capacity: 1.00
Maximum capacity: 1.00
Application count: 1
----- Applications in this queue -----
Application ID: application_1431052132967_0001 (H2O_3889)
Started: root (Fri May 08 02:33:50 UTC 2015)
Application state: FINISHED
Tracking URL: http://sandbox.hortonworks.com:8088/proxy/application_1431052132967_0001/jobhistory/job/job_1431052132967_0001
Queue name: default
Used/Reserved containers: 1 / 0
Needed/Used/Reserved memory: 0.2 GB / 0.2 GB / 0.0 GB
Needed/Used/Reserved vcores: 1 / 1 / 0
Queue 'default' approximate utilization: 0.2 / 2.2 GB used, 1 / 8 vcores used
----------------------------------------------------------------------
ERROR: Job memory request (2.2 GB) exceeds available YARN cluster memory (2.2 GB)
WARNING: Job memory request (2.2 GB) exceeds queue available memory capacity (2.0 GB)
ERROR: Only 1 out of the requested 2 worker containers were started due to YARN cluster resource limitations
----------------------------------------------------------------------
Attempting to clean up hadoop job...
15/05/08 02:35:59 INFO impl.YarnClientImpl: Killed application application_1431052132967_0001
Killed.
[root@sandbox h2o-dev-0.3.0.1188-hdp2.2]#
The H2O launch failed because more memory was requested than was available. Make sure you are not trying to specify more memory in the launch parameters than you have available.
How does the architecture of H2O work?
This PDF includes diagrams and slides depicting how H2O works in big data environments.
I received the following error message when launching H2O - how do I resolve the error?
Invalid flow_dir illegal character at index 12...
This error message means that there is a space (or other unsupported character) in your H2O directory. To resolve this error:
Create a new folder without unsupported characters to use as the H2O directory (for example,
C:\h2o).or
Specify a different save directory using the
-flow_dirparameter when launching H2O:java -jar h2o.jar -flow_dir test
How does importFiles() work in H2O?
importFiles() gets the basic information for the file and then returns a key representing that file. This key is used during parsing to read in the file and to save space so that the file isn’t loaded every time; instead, it is loaded into H2O then referenced using the key. For files hosted online, H2O verifies the destination is valid, creates a vec that loads the file when necessary, and returns a key.
Does H2O support GPUs?
Currently, we do not support this capability. If you are interested in contributing your efforts to support this feature to our open-source code database, please contact us at h2ostream@googlegroups.com.
How can I continue working on a model in H2O after restarting?
There are a number of ways you can save your model in H2O:
- In the web UI, click the Flow menu then click Save Flow. Your flow is saved to the Flows tab in the Help sidebar on the right.
- In the web UI, click the Flow menu then click Download this Flow…. Depending on your browser and configuration, your flow is saved to the “Downloads” folder (by default) or to the location you specify in the pop-up Save As window if it appears.
- (For DRF, GBM, and DL models only): Use model checkpointing to resume training a model. Copy the
model_idnumber from a built model and paste it into the checkpoint field in thebuildModelcell.
How can I find out more about H2O’s real-time, nano-fast scoring engine?
H2O’s scoring engine uses a Plain Old Java Object (POJO). The POJO code runs quickly but is single-threaded. It is intended for embedding into lightweight real-time environments.
All the work is done by the call to the appropriate predict method. There is no involvement from H2O in this case.
To compare multiple models simultaneously, use the POJO to call the models using multiple threads. For more information on using POJOs, refer to the POJO Quick Start Guide and POJO Java Documentation
In-H2O scoring is triggered on an existing H2O cluster, typically using a REST API call. H2O evaluates the predictions in a parallel and distributed fashion for this case. The predictions are stored into a new Frame and can be written out using h2o.exportFile(), for example.
I am using an older version of H2O (2.8 or prior) - where can I find documentation for this version?
If you are using H2O 2.8 or prior, we strongly recommend upgrading to the latest version of H2O if possible.
If you do not wish to upgrade to the latest version, documentation for H2O Classic is available here.
I am writing an academic research paper and I would like to cite H2O in my bibliography - how should I do that?
To cite our software:
The H2O.ai Team. (2015) h2o: R Interface for H2O. R package version 3.1.0.99999. http://www.h2o.ai.
The H2O.ai Team. (2015) h2o: h2o: Python Interface for H2O. Python package version 3.1.0.99999. http://www.h2o.ai.
- The H2O.ai Team. (2015) H2O: Scalable Machine Learning. Version 3.1.0.99999. http://www.h2o.ai.
To cite one of our booklets:
Nykodym, T., Hussami, N., Kraljevic, T.,Rao, A., and Wang, A. (Sept. 2015). Generalized Linear Modeling with H2O. http://h2o.ai/resources.
Candel, A., LeDell, E., Parmar, V., and Arora, A. (Sept. 2015). Deep Learning with H2O. http://h2o.ai/resources.
Click, C., Malohlava, M., Parmar, V., and Roark, H. (Sept. 2015). Gradient Boosted Models with H2O. http://h2o.ai/resources.
Aiello, S., Eckstrand, E., Fu, A., Landry, M., and Aboyoun, P. (Sept. 2015) Fast Scalable R with H2O. http://h2o.ai/resources.
Aiello, S., Click, C., Roark, H. and Rehak, L. (Sept. 2015) Machine Learning with Python and H2O http://h2o.ai/resources.
Malohlava, M., and Tellez, A. (Sept. 2015) Machine Learning with Sparkling Water: H2O + Spark http://h2o.ai/resources.
If you are using Bibtex:
@Manual{h2o_GLM_booklet,
title = {Generalized Linear Modeling with H2O},
author = {Nykodym, T. and Hussami, N. and Kraljevic, T. and Rao, A. and Wang, A.},
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_DL_booklet,
title = {Deep Learning with H2O},
author = {Candel, A. and LeDell, E. and Arora, A. and Parmar, V.},
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_GBM_booklet,
title = {Gradient Boosted Models},
author = {Click, C. and Lanford, J. and Malohlava, M. and Parmar, V. and Roark, H.},
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_R_booklet,
title = {Fast Scalable R with H2O},
author = {Aiello, S. and Eckstrand, E. and Fu, A. and Landry, M. and Aboyoun, P. },
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_R_package,
title = {h2o: R Interface for H2O},
author = {The H2O.ai team},
year = {2015},
note = {R package version 3.1.0.99999},
url = {http://www.h2o.ai},
}
@Manual{h2o_Python_module,
title = {h2o: Python Interface for H2O},
author = {The H2O.ai team},
year = {2015},
note = {Python package version 3.1.0.99999},
url = {http://www.h2o.ai},
}
@Manual{h2o_Java_software,
title = {H2O: Scalable Machine Learning},
author = {The H2O.ai team},
year = {2015},
note = {version 3.1.0.99999},
url = {http://www.h2o.ai},
}
How can I use Flow to export the prediction results with a dataset?
After obtaining your results, click the Combine predictions with frame button, then click the View Frame button.
What are these RTMP and py_ temporary Frames? Why are they the same size as my original data?
No data is copied. H2O does a classic copy-on-write optimization. That Frame you see - it’s nothing more than a thin wrapper over an internal list of columns; the columns are shared to avoid the copying.
The RTMP’s now need to be entirely managed by the H2O wrapper - because indeed they are using shared state under the hood. If you delete one, you probably delete parts of others. Instead, temp management should be automatic and “good” - as in: it’s a bug if you need to delete a temp manually, or if passing around Frames, or adding or removing columns turns into large data copies.
R’s GC is now used to remove unused R temps, and when the last use of a shared column goes away, then the H2O wrapper will tell the H2O cluster to remove that no longer needed column.
In other words: Don’t delete RTMPs, they’ll disappear at the next R GC. Don’t worry about copies (they aren’t getting made). Do Nothing and All Is Well.
Hadoop
Why did I get an error in R when I tried to save my model to my home directory in Hadoop?
To save the model in HDFS, prepend the save directory with hdfs://:
# build model
model = h2o.glm(model params)
# save model
hdfs_name_node <- "mr-0x6"
hdfs_tmp_dir <- "/tmp/runit”
model_path <- sprintf("hdfs://%s%s", hdfs_name_node, hdfs_tmp_dir)
h2o.saveModel(model, dir = model_path, name = “mymodel")
How do I specify which nodes should run H2O in a Hadoop cluster?
After creating and applying the desired node labels and associating them with specific queues as described in the Hadoop documentation, launch H2O using the following command:
hadoop jar h2odriver.jar -Dmapreduce.job.queuename=<my-h2o-queue> -nodes <num-nodes> -mapperXmx 6g -output hdfsOutputDirName
-Dmapreduce.job.queuename=<my-h2o-queue>represents the queue name-nodes <num-nodes>represents the number of nodes-mapperXmx 6glaunches H2O with 6g of memory-output hdfsOutputDirNamespecifies the HDFS output directory ashdfsOutputDirName
How do I import data from HDFS in R and in Flow?
To import from HDFS in R:
h2o.importFolder(path, pattern = "", destination_frame = "", parse = TRUE, header = NA, sep = "", col.names = NULL, na.strings = NULL)
Here is another example:
# pathToAirlines <- "hdfs://mr-0xd6.0xdata.loc/datasets/airlines_all.csv"
# airlines.hex <- h2o.importFile(path = pathToAirlines, destination_frame = "airlines.hex")
In Flow, the easiest way is to let the auto-suggestion feature in the Search: field complete the path for you. Just start typing the path to the file, starting with the top-level directory, and H2O provides a list of matching files.

Click the file to add it to the Search: field.
Why do I receive the following error when I try to save my notebook in Flow?
Error saving notebook: Error calling POST /3/NodePersistentStorage/notebook/Test%201 with opts
When you are running H2O on Hadoop, H2O tries to determine the home HDFS directory so it can use that as the download location. If the default home HDFS directory is not found, manually set the download location from the command line using the -flow_dir parameter (for example, hadoop jar h2odriver.jar <...> -flow_dir hdfs:///user/yourname/yourflowdir). You can view the default download directory in the logs by clicking Admin > View logs… and looking for the line that begins Flow dir:.
How do I access data in HDFS without launching H2O on Yarn?
Each h2odriver.jar file is built with a specific Hadoop distribution so in order to have a working HDFS connection download the h2odriver.jar file for your Hadoop distribution.
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-cdh5.2.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-cdh5.3.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-hdp2.1.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-hdp2.2.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-hdp2.3.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-mapr3.1.1.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-mapr4.0.1.zip
wget http://h2o-release.s3.amazonaws.com/h2o/master/3/h2o-3.8.2.3-mapr5.0.zip
**Note**: Enter only one of the above commands.
Then run the command to launch the H2O Application in the driver by specifying the classpath:
unzip h2o-3.8.2.3-*.zip
cd h2o-3.8.2.3-*
java -cp h2odriver.jar water.H2OApp
Java
How do I use H2O with Java?
There are two ways to use H2O with Java. The simplest way is to call the REST API from your Java program to a remote cluster and should meet the needs of most users.
You can access the REST API documentation within Flow, or on our documentation site.
Flow, Python, and R all rely on the REST API to run H2O. For example, each action in Flow translates into one or more REST API calls. The script fragments in the cells in Flow are essentially the payloads for the REST API calls. Most R and Python API calls translate into a single REST API call.
To see how the REST API is used with H2O:
Using Chrome as your internet browser, open the developer tab while viewing the web UI. As you perform tasks, review the network calls made by Flow.
Write an R program for H2O using the H2O R package that uses
h2o.startLogging()at the beginning. All REST API calls used are logged.
The second way to use H2O with Java is to embed H2O within your Java application, similar to Sparkling Water.
How do I communicate with a remote cluster using the REST API?
To create a set of bare POJOs for the REST API payloads that can be used by JVM REST API clients:
- Clone the sources from GitHub.
- Start an H2O instance.
- Enter
% cd py. - Enter
% python generate_java_binding.py.
This script connects to the server, gets all the metadata for the REST API schemas, and writes the Java POJOs to {sourcehome}/build/bindings/Java.
I keep getting a message that I need to install Java. I have the latest version of Java installed, but I am still getting this message. What should I do?
This error message displays if the JAVA_HOME environment variable is not set correctly. The JAVA_HOME variable is likely points to Apple Java version 6 instead of Oracle Java version 8.
If you are running OS X 10.7 or earlier, enter the following in Terminal:
export JAVA_HOME=/Library/Internet\ Plug-Ins/JavaAppletPlugin.plugin/Contents/Home
If you are running OS X 10.8 or later, modify the launchd.plist by entering the following in Terminal:
cat << EOF | sudo tee /Library/LaunchDaemons/setenv.JAVA_HOME.plist
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE plist PUBLIC "-//Apple//DTD PLIST 1.0//EN" "http://www.apple.com/DTDs/PropertyList-1.0.dtd">
<plist version="1.0">
<dict>
<key>Label</key>
<string>setenv.JAVA_HOME</string>
<key>ProgramArguments</key>
<array>
<string>/bin/launchctl</string>
<string>setenv</string>
<string>JAVA_HOME</string>
<string>/Library/Internet Plug-Ins/JavaAppletPlugin.plugin/Contents/Home</string>
</array>
<key>RunAtLoad</key>
<true/>
<key>ServiceIPC</key>
<false/>
</dict>
</plist>
EOF
Python
I tried to install H2O in Python but pip install scikit-learn failed - what should I do?
Use the following commands (prepending with sudo if necessary):
easy_install pip
pip install numpy
brew install gcc
pip install scipy
pip install scikit-learn
If you are still encountering errors and you are using OSX, the default version of Python may be installed. We recommend installing the Homebrew version of Python instead:
brew install python
If you are encountering errors related to missing Python packages when using H2O, refer to the following list for a complete list of all Python packages, including dependencies:
grip |
tabulate |
wheel |
jsonlite |
ipython |
numpy |
scipy |
pandas |
-U gensim |
jupyter |
-U PIL |
nltk |
beautifulsoup4 |
|
|
How do I specify a value as an enum in Python? Is there a Python equivalent of as.factor() in R?
Use .asfactor() to specify a value as an enum.
I received the following error when I tried to install H2O using the Python instructions on the downloads page - what should I do to resolve it?
Downloading/unpacking http://h2o-release.s3.amazonaws.com/h2o/rel-shannon/12/Python/h2o-3.0.0.12-py2.py3-none-any.whl
Downloading h2o-3.0.0.12-py2.py3-none-any.whl (43.1Mb): 43.1Mb downloaded
Running setup.py egg_info for package from http://h2o-release.s3.amazonaws.com/h2o/rel-shannon/12/Python/h2o-3.0.0.12-py2.py3-none-any.whl
Traceback (most recent call last):
File "<string>", line 14, in <module>
IOError: [Errno 2] No such file or directory: '/tmp/pip-nTu3HK-build/setup.py'
Complete output from command python setup.py egg_info:
Traceback (most recent call last):
File "<string>", line 14, in <module>
IOError: [Errno 2] No such file or directory: '/tmp/pip-nTu3HK-build/setup.py'
---
Command python setup.py egg_info failed with error code 1 in /tmp/pip-nTu3HK-build
With Python, there is no automatic update of installed packages, so you must upgrade manually. Additionally, the package distribution method recently changed from distutils to wheel. The following procedure should be tried first if you are having trouble installing the H2O package, particularly if error messages related to bdist_wheel or eggs display.
# this gets the latest setuptools
# see https://pip.pypa.io/en/latest/installing.html
wget https://bootstrap.pypa.io/ez_setup.py -O - | sudo python
# platform dependent ways of installing pip are at
# https://pip.pypa.io/en/latest/installing.html
# but the above should work on most linux platforms?
# on ubuntu
# if you already have some version of pip, you can skip this.
sudo apt-get install python-pip
# the package manager doesn't install the latest. upgrade to latest
# we're not using easy_install any more, so don't care about checking that
pip install pip --upgrade
# I've seen pip not install to the final version ..i.e. it goes to an almost
# final version first, then another upgrade gets it to the final version.
# We'll cover that, and also double check the install.
# after upgrading pip, the path name may change from /usr/bin to /usr/local/bin
# start a new shell, just to make sure you see any path changes
bash
# Also: I like double checking that the install is bulletproof by reinstalling.
# Sometimes it seems like things say they are installed, but have errors during the install. Check for no errors or stack traces.
pip install pip --upgrade --force-reinstall
# distribute should be at the most recent now. Just in case
# don't do --force-reinstall here, it causes an issue.
pip install distribute --upgrade
# Now check the versions
pip list | egrep '(distribute|pip|setuptools)'
distribute (0.7.3)
pip (7.0.3)
setuptools (17.0)
# Re-install wheel
pip install wheel --upgrade --force-reinstall
After completing this procedure, go to Python and use h2o.init() to start H2O in Python.
Note:
If you use gradlew to build the jar yourself, you have to start the jar >yourself before you do
h2o.init().If you download the jar and the H2O package,
h2o.init()will work like R >and you don’t have to start the jar yourself.
How should I specify the datatype during import in Python?
Refer to the following example:
#Let's say you want to change the second column "CAPSULE" of prostate.csv
#to categorical. You have 3 options.
#Option 1. Use a dictionary of column names to types.
fr = h2o.import_file("smalldata/logreg/prostate.csv", col_types = {"CAPSULE":"Enum"})
fr.describe()
#Option 2. Use a list of column types.
c_types = [None]*9
c_types[1] = "Enum"
fr = h2o.import_file("smalldata/logreg/prostate.csv", col_types = c_types)
fr.describe()
#Option 3. Use parse_setup().
fraw = h2o.import_file("smalldata/logreg/prostate.csv", parse = False)
fsetup = h2o.parse_setup(fraw)
fsetup["column_types"][1] = '"Enum"'
fr = h2o.parse_raw(fsetup)
fr.describe()
How do I view a list of variable importances in Python?
Use model.varimp(return_list=True) as shown in the following example:
model = h2o.gbm(y = "IsDepDelayed", x = ["Month"], training_frame = df)
vi = model.varimp(return_list=True)
Out[26]:
[(u'Month', 69.27436828613281, 1.0, 1.0)]
What is PySparkling? How can I use it for grid search or early stopping?
PySparkling basically calls H2O Python functions for all operations on H2O data frames. You can perform all H2O Python operations available in H2O Python version 3.6.0.3 or later from PySparkling.
For help on a function within IPython Notebook, run H2OGridSearch?
Here is an example of grid search in PySparkling:
from h2o.grid.grid_search import H2OGridSearch
from h2o.estimators.gbm import H2OGradientBoostingEstimator
iris = h2o.import_file("/Users/nidhimehta/h2o-dev/smalldata/iris/iris.csv")
ntrees_opt = [5, 10, 15]
max_depth_opt = [2, 3, 4]
learn_rate_opt = [0.1, 0.2]
hyper_parameters = {"ntrees": ntrees_opt, "max_depth":max_depth_opt,
"learn_rate":learn_rate_opt}
gs = H2OGridSearch(H2OGradientBoostingEstimator(distribution='multinomial'), hyper_parameters)
gs.train(x=range(0,iris.ncol-1), y=iris.ncol-1, training_frame=iris, nfold=10)
#gs.show
print gs.sort_by('logloss', increasing=True)
Here is an example of early stopping in PySparkling:
from h2o.grid.grid_search import H2OGridSearch
from h2o.estimators.deeplearning import H2ODeepLearningEstimator
hidden_opt = [[32,32],[32,16,8],[100]]
l1_opt = [1e-4,1e-3]
hyper_parameters = {"hidden":hidden_opt, "l1":l1_opt}
model_grid = H2OGridSearch(H2ODeepLearningEstimator, hyper_params=hyper_parameters)
model_grid.train(x=x, y=y, distribution="multinomial", epochs=1000, training_frame=train,
validation_frame=test, score_interval=2, stopping_rounds=3, stopping_tolerance=0.05, stopping_metric="misclassification")
Do you have a tutorial for grid search in Python?
Yes, a notebook is available here that demonstrates the use of grid search in Python.
R
Which versions of R are compatible with H2O?
Currently, the only version of R that is known to not work well with H2O is R version 3.1.0 (codename “Spring Dance”). If you are using this version, we recommend upgrading the R version before using H2O.
What R packages are required to use H2O?
The following packages are required:
methodsstatmodstatsgraphicsRCurljsonlitetoolsutils
Some of these packages have dependencies; for example, bitops is required, but it is a dependency of the RCurl package, so bitops is automatically included when RCurl is installed.
If you are encountering errors related to missing R packages when using H2O, refer to the following list for a complete list of all R packages, including dependencies:
statmod |
bitops |
RCurl |
jsonlite |
methods |
stats |
graphics |
tools |
utils |
stringi |
magrittr |
colorspace |
stringr |
RColorBrewer |
dichromat |
munsell |
labeling |
plyr |
digest |
gtable |
reshape2 |
scales |
proto |
ggplot2 |
h2oEnsemble |
gtools |
gdata |
caTools |
gplots |
chron |
ROCR |
data.table |
cvAUC |
How can I install the H2O R package if I am having permissions problems?
This issue typically occurs for Linux users when the R software was installed by a root user. For more information, refer to the following link.
To specify the installation location for the R packages, create a file that contains the R_LIBS_USER environment variable:
echo R_LIBS_USER=\"~/.Rlibrary\" > ~/.Renviron
Confirm the file was created successfully using cat:
$ cat ~/.Renviron
You should see the following output:
R_LIBS_USER="~/.Rlibrary"
Create a new directory for the environment variable:
$ mkdir ~/.Rlibrary
Start R and enter the following:
.libPaths()
Look for the following output to confirm the changes:
[1] "<Your home directory>/.Rlibrary"
[2] "/Library/Frameworks/R.framework/Versions/3.1/Resources/library"
I received the following error message after launching H2O in RStudio and using h2o.init - what should I do to resolve this error?
Error in h2o.init() :
Version mismatch! H2O is running version 3.2.0.9 but R package is version 3.2.0.3
This error is due to a version mismatch between the H2O R package and the running H2O instance. Make sure you are using the latest version of both files by downloading H2O from the downloads page and installing the latest version and that you have removed any previous H2O R package versions by running:
if ("package:h2o" %in% search()) { detach("package:h2o", unload=TRUE) }
if ("h2o" %in% rownames(installed.packages())) { remove.packages("h2o") }
Make sure to install the dependencies for the H2O R package as well:
if (! ("methods" %in% rownames(installed.packages()))) { install.packages("methods") }
if (! ("statmod" %in% rownames(installed.packages()))) { install.packages("statmod") }
if (! ("stats" %in% rownames(installed.packages()))) { install.packages("stats") }
if (! ("graphics" %in% rownames(installed.packages()))) { install.packages("graphics") }
if (! ("RCurl" %in% rownames(installed.packages()))) { install.packages("RCurl") }
if (! ("jsonlite" %in% rownames(installed.packages()))) { install.packages("jsonlite") }
if (! ("tools" %in% rownames(installed.packages()))) { install.packages("tools") }
if (! ("utils" %in% rownames(installed.packages()))) { install.packages("utils") }
Finally, install the latest version of the H2O package for R:
install.packages("h2o", type="source", repos=(c("http://h2o-release.s3.amazonaws.com/h2o/master/3/R")))
library(h2o)
localH2O = h2o.init(nthreads=-1)
If your R version is older than the H2O R package, upgrade your R version using update.packages(checkBuilt=TRUE, ask=FALSE).
I received the following error message after trying to run some code - what should I do?
> fit <- h2o.deeplearning(x=2:4, y=1, training_frame=train_hex)
|=========================================================================================================| 100%
Error in model$training_metrics$MSE :
$ operator not defined for this S4 class
In addition: Warning message:
Not all shim outputs are fully supported, please see ?h2o.shim for more information
Remove the h2o.shim(enable=TRUE) line and try running the code again. Note that the h2o.shim is only a way to notify users of previous versions of H2O about changes to the H2O R package - it will not revise your code, but provides suggested replacements for deprecated commands and parameters.
How do I extract the model weights from a model I’ve creating using H2O in R? I’ve enabled extract_model_weights_and_biases, but the output refers to a file I can’t open in R.
For an example of how to extract weights and biases from a model, refer to the following repo location on GitHub.
How do I extract the run time of my model as output?
For the following example:
out.h2o.rf = h2o.randomForest( x=c("x1", "x2", "x3", "w"), y="y", training_frame=h2o.df.train, seed=555, model_id= "my.model.1st.try.out.h2o.rf" )
Use out.h2o.rf@model$run_time to determine the value of the run_time variable.
What is the best way to do group summarizations? For example, getting sums of specific columns grouped by a categorical column.
We strongly recommend using h2o.group_by for this function instead of h2o.ddply, as shown in the following example:
newframe <- h2o.group_by(h2oframe, by="footwear_category", nrow("email_event_click_ct"), sum("email_event_click_ct"), mean("email_event_click_ct"), sd("email_event_click_ct"), gb.control = list( col.names=c("count", "total_email_event_click_ct", "avg_email_event_click_ct", "std_email_event_click_ct") ) )
Using gb.control is optional; here it is included so the column names are user-configurable.
The by option can take a list of columns if you want to group by more than one column to compute the summary as shown in the following example:
newframe <- h2o.group_by(h2oframe, by=c("footwear_category","age_group"), nrow("email_event_click_ct"), sum("email_event_click_ct"), mean("email_event_click_ct"), sd("email_event_click_ct"), gb.control = list( col.names=c("count", "total_email_event_click_ct", "avg_email_event_click_ct", "std_email_event_click_ct") ) )
I’m using CentOS and I want to run H2O in R - are there any dependencies I need to install?
Yes, make sure to install libcurl, which allows H2O to communicate with R. We also recommend disabling SElinux and any firewalls, at least initially until you have confirmed H2O can initialize.
How do I change variable/header names on an H2O frame in R?
There are two ways to change header names. To specify the headers during parsing, import the headers in R and then specify the header as the column name when the actual data frame is imported:
header <- h2o.importFile(path = pathToHeader)
data <- h2o.importFile(path = pathToData, col.names = header)
data
You can also use the names() function:
header <- c("user", "specified", "column", "names")
data <- h2o.importFile(path = pathToData)
names(data) <- header
To replace specific column names, you can also use a sub/gsub in R:
header <- c("user", "specified", "column", "names")
To replace “user” column with “computer”
data <- h2o.importFile(path = pathToData)
names(data) <- sub(pattern = "user", replacement = "computer", x = names(header))
My R terminal crashed - how can I re-access my H2O frame?
Launch H2O and use your web browser to access the web UI, Flow, at localhost:54321. Click the Data menu, then click List All Frames. Copy the frame ID, then run h2o.ls() in R to list all the frames, or use the frame ID in the following code (replacing YOUR_FRAME_ID with the frame ID):
library(h2o)
localH2O = h2o.init(ip="sri.h2o.ai", port=54321, startH2O = F, strict_version_check=T)
data_frame <- h2o.getFrame(frame_id = "YOUR_FRAME_ID")
How do I remove rows containing NAs in an H2OFrame?
To remove NAs from rows:
a b c d e
1 0 NA NA NA NA
2 0 2 2 2 2
3 0 NA NA NA NA
4 0 NA NA 1 2
5 0 NA NA NA NA
6 0 1 2 3 2
Removing rows 1, 3, 4, 5 to get:
a b c d e
2 0 2 2 2 2
6 0 1 2 3 2
Use na.omit(myFrame), where myFrame represents the name of the frame you are editing.
I installed H2O in R using OS X and updated all the dependencies, but the following error message displayed: Error in .h2o.doSafeREST(h2oRestApiVersion = h2oRestApiVersion, Unexpected CURL error: Empty reply from server - what should I do?
This error message displays if the JAVA_HOME environment variable is not set correctly. The JAVA_HOME variable is likely points to Apple Java version 6 instead of Oracle Java version 8.
If you are running OS X 10.7 or earlier, enter the following in Terminal:
export JAVA_HOME=/Library/Internet\ Plug-Ins/JavaAppletPlugin.plugin/Contents/Home
If you are running OS X 10.8 or later, modify the launchd.plist by entering the following in Terminal:
cat << EOF | sudo tee /Library/LaunchDaemons/setenv.JAVA_HOME.plist
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE plist PUBLIC "-//Apple//DTD PLIST 1.0//EN" "http://www.apple.com/DTDs/PropertyList-1.0.dtd">
<plist version="1.0">
<dict>
<key>Label</key>
<string>setenv.JAVA_HOME</string>
<key>ProgramArguments</key>
<array>
<string>/bin/launchctl</string>
<string>setenv</string>
<string>JAVA_HOME</string>
<string>/Library/Internet Plug-Ins/JavaAppletPlugin.plugin/Contents/Home</string>
</array>
<key>RunAtLoad</key>
<true/>
<key>ServiceIPC</key>
<false/>
</dict>
</plist>
EOF
How does the col.names argument work in group_by?
You need to add the col.names inside the gb.control list. Refer to the following example:
newframe <- h2o.group_by(dd, by="footwear_category", nrow("email_event_click_ct"), sum("email_event_click_ct"), mean("email_event_click_ct"),
sd("email_event_click_ct"), gb.control = list( col.names=c("count", "total_email_event_click_ct", "avg_email_event_click_ct", "std_email_event_click_ct") ) )
newframe$avg_email_event_click_ct2 = newframe$total_email_event_click_ct / newframe$count
How are the results of h2o.predict displayed?
The order of the rows in the results for h2o.predict is the same as the order in which the data was loaded, even if some rows fail (for example, due to missing values or unseen factor levels). To bind a per-row identifier, use cbind.
How do I view all the variable importances for a model?
By default, H2O returns the top five and lowest five variable importances. To view all the variable importances, use the following:
model <- h2o.getModel(model_id = "my_H2O_modelID",conn=localH2O)
varimp<-as.data.frame(h2o.varimp(model))
How do I add random noise to a column in an H2O frame?
To add random noise to a column in an H2O frame, refer to the following example:
h2o.init()
fr <- as.h2o(iris)
|======================================================================| 100%
random_column <- h2o.runif(fr)
new_fr <- h2o.cbind(fr,random_column)
new_fr
Sparkling Water
What are the advantages of using Sparkling Water compared with H2O?
Sparkling Water contains the same features and functionality as H2O but provides a way to use H2O with Spark, a large-scale cluster framework.
Sparkling Water is ideal for H2O users who need to manage large clusters for their data processing needs and want to transfer data from Spark to H2O (or vice versa).
There is also a Python interface available to enable access to Sparkling Water directly from PySpark.
How do I filter an H2OFrame using Sparkling Water?
Filtering columns is easy: just remove the unnecessary columns or create a new H2OFrame from the columns you want to include (Frame(String[] names, Vec[] vec)), then make the H2OFrame wrapper around it (new H2OFrame(frame)).
Filtering rows is a little bit harder. There are two ways:
Create an additional binary vector holding
1/0for thein/outsample (make sure to take this additional vector into account in your computations). This solution is quite cheap, since you do not duplicate data - just create a simple vector in a data walk.or
Create a new frame with the filtered rows. This is a harder task, since you have to copy data. For reference, look at the #deepSlice call on Frame (
H2OFrame)
How can I save and load a K-means model using Sparkling Water?
The following example code defines the save and load functions explicitly.
import water._
import _root_.hex._
import java.net.URI
import water.serial.ObjectTreeBinarySerializer
// Save H2O model (as binary)
def exportH2OModel(model : Model[_,_,_], destination: URI): URI = {
val modelKey = model._key.asInstanceOf[Key[_ <: Keyed[_ <: Keyed[_ <: AnyRef]]]]
val keysToExport = model.getPublishedKeys()
// Prepend model key
keysToExport.add(0, modelKey)
new ObjectTreeBinarySerializer().save(keysToExport, destination)
destination
}
// Get model from H2O DKV and Save to disk
val gbmModel: _root_.hex.tree.gbm.GBMModel = DKV.getGet("model")
exportH2OModel(gbmModel, new File("../h2omodel.bin").toURI)
def loadH2OModel[M <: Model[_, _, _]](source: URI) : M = {
val l = new ObjectTreeBinarySerializer().load(source)
l.get(0).get().asInstanceOf[M]
}
// Load H2O model
def loadH2OModel[M <: Model[_, _, _]](source: URI) : M = {
val l = new ObjectTreeBinarySerializer().load(source)
l.get(0).get().asInstanceOf[M]
}
// Load model
val h2oModel: Model[_, _, _] = loadH2OModel(new File("../h2omodel.bin").toURI)
How do I inspect H2O using Flow while a droplet is running?
If your droplet execution time is very short, add a simple sleep statement to your code:
Thread.sleep(...)
How do I change the memory size of the executors in a droplet?
There are two ways to do this:
Change your default Spark setup in
$SPARK_HOME/conf/spark-defaults.confor
Pass
--confvia spark-submit when you launch your droplet (e.g.,$SPARK_HOME/bin/spark-submit --conf spark.executor.memory=4g --master $MASTER --class org.my.Droplet $TOPDIR/assembly/build/libs/droplet.jar
I received the following error while running Sparkling Water using multiple nodes, but not when using a single node - what should I do?
onExCompletion for water.parser.ParseDataset$MultiFileParseTask@31cd4150
water.DException$DistributedException: from /10.23.36.177:54321; by class water.parser.ParseDataset$MultiFileParseTask; class water.DException$DistributedException: from /10.23.36.177:54325; by class water.parser.ParseDataset$MultiFileParseTask; class water.DException$DistributedException: from /10.23.36.178:54325; by class water.parser.ParseDataset$MultiFileParseTask$DistributedParse; class java.lang.NullPointerException: null
at water.persist.PersistManager.load(PersistManager.java:141)
at water.Value.loadPersist(Value.java:226)
at water.Value.memOrLoad(Value.java:123)
at water.Value.get(Value.java:137)
at water.fvec.Vec.chunkForChunkIdx(Vec.java:794)
at water.fvec.ByteVec.chunkForChunkIdx(ByteVec.java:18)
at water.fvec.ByteVec.chunkForChunkIdx(ByteVec.java:14)
at water.MRTask.compute2(MRTask.java:426)
at water.MRTask.compute2(MRTask.java:398)
This error output displays if the input file is not present on all nodes. Because of the way that Sparkling Water distributes data, the input file is required on all nodes (including remote), not just the primary node. Make sure there is a copy of the input file on all the nodes, then try again.
Are there any drawbacks to using Sparkling Water compared to standalone H2O?
The version of H2O embedded in Sparkling Water is the same as the standalone version.
How do I use Sparkling Water from the Spark shell?
There are two methods:
Use
$SPARK_HOME/bin/spark-shell --packages ai.h2o:sparkling-water-core_2.10:1.3.3or
bin/sparkling-shell
The software distribution provides example scripts in the examples/scripts directory:
bin/sparkling-shell -i examples/scripts/chicagoCrimeSmallShell.script.scala
For either method, initialize H2O as shown in the following example:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc).start()
After successfully launching H2O, the following output displays:
Sparkling Water Context:
* number of executors: 3
* list of used executors:
(executorId, host, port)
------------------------
(1,Michals-MBP.0xdata.loc,54325)
(0,Michals-MBP.0xdata.loc,54321)
(2,Michals-MBP.0xdata.loc,54323)
------------------------
Open H2O Flow in browser: http://172.16.2.223:54327 (CMD + click in Mac OSX)
How do I use H2O with Spark Submit?
Spark Submit is for submitting self-contained applications. For more information, refer to the Spark documentation.
First, initialize H2O:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc).start()
The Sparkling Water distribution provides several examples of self-contained applications built with Sparkling Water. To run the examples:
bin/run-example.sh ChicagoCrimeAppSmall
The “magic” behind run-example.sh is a regular Spark Submit:
$SPARK_HOME/bin/spark-submit ChicagoCrimeAppSmall --packages ai.h2o:sparkling-water-core_2.10:1.3.3 --packages ai.h2o:sparkling-water-examples_2.10:1.3.3
How do I use Sparkling Water with Databricks cloud?
Sparkling Water compatibility with Databricks cloud is still in development.
How do I develop applications with Sparkling Water?
For a regular Spark application (a self-contained application as described in the Spark documentation), the app needs to initialize H2OServices via H2OContext:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc).start()
For more information, refer to the Sparkling Water development documentation.
How do I connect to Sparkling Water from R or Python?
After starting H2OServices by starting H2OContext, point your client to the IP address and port number specified in H2OContext.
I’m getting a java.lang.ArrayIndexOutOfBoundsException when I try to run Sparkling Water - what do I need to do to resolve this error?
This error message displays if you have not set up the H2OContext before running Sparkling Water. To set up the H2OContext:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc)
After setting up H2OContext, try to run Sparkling Water again.
Tunneling between servers with H2O
To tunnel between servers (for example, due to firewalls):
- Use ssh to log in to the machine where H2O will run.
Start an instance of H2O by locating the working directory and calling a java command similar to the following example.
The port number chosen here is arbitrary; yours may be different.
$ java -jar h2o.jar -port 55599This returns output similar to the following:
irene@mr-0x3:~/target$ java -jar h2o.jar -port 55599 04:48:58.053 main INFO WATER: ----- H2O started ----- 04:48:58.055 main INFO WATER: Build git branch: master 04:48:58.055 main INFO WATER: Build git hash: 64fe68c59ced5875ac6bac26a784ce210ef9f7a0 04:48:58.055 main INFO WATER: Build git describe: 64fe68c 04:48:58.055 main INFO WATER: Build project version: 1.7.0.99999 04:48:58.055 main INFO WATER: Built by: 'Irene' 04:48:58.055 main INFO WATER: Built on: 'Wed Sep 4 07:30:45 PDT 2013' 04:48:58.055 main INFO WATER: Java availableProcessors: 4 04:48:58.059 main INFO WATER: Java heap totalMemory: 0.47 gb 04:48:58.059 main INFO WATER: Java heap maxMemory: 6.96 gb 04:48:58.060 main INFO WATER: ICE root: '/tmp' 04:48:58.081 main INFO WATER: Internal communication uses port: 55600 + Listening for HTTP and REST traffic on + http://192.168.1.173:55599/ 04:48:58.109 main INFO WATER: H2O cloud name: 'irene' 04:48:58.109 main INFO WATER: (v1.7.0.99999) 'irene' on /192.168.1.173:55599, discovery address /230 .252.255.19:59132 04:48:58.111 main INFO WATER: Cloud of size 1 formed [/192.168.1.173:55599] 04:48:58.247 main INFO WATER: Log dir: '/tmp/h2ologs'Log into the remote machine where the running instance of H2O will be forwarded using a command similar to the following (your specified port numbers and IP address will be different)
ssh -L 55577:localhost:55599 irene@192.168.1.173Check the cluster status.
You are now using H2O from localhost:55577, but the instance of H2O is running on the remote server (in this case the server with the ip address 192.168.1.xxx) at port number 55599.
To see this in action note that the web UI is pointed at localhost:55577, but that the cluster status shows the cluster running on 192.168.1.173:55599
Glossary
| Term | Definition | |
|---|---|---|
| H2O.ai | Maker of H2O. Visit our website. | |
| Autoencoder | An extension of the Deep Learning framework. Can be used to compress input features (similar to PCA). Sparse autoencoders are simple extensions that can increase accuracy. Use autoencoders for: - generic dimensionality reduction (for pre-processing for any algorithm) - anomaly detection (for comparing the reconstructed signal with the original to find differences that may be anomalies) - layer-by-layer pre-training (using stacked auto-encoders) |
|
| Backpropogation | Uses a known, desired output for each input value to calculate the loss function gradient for training. If enabled, performed after each training sample in Deep Learning. | |
| BAD | A column type that contains only missing values. | |
| Balance classes | A parameter that oversamples the minority classes to balance the distribution. | |
| Beta constraints | A data.frame or H2OParsedData object with the columns [“names”, “lower_bounds”,”upper_bounds”, “beta_given”], where each row corresponds to a predictor in the GLM. “names” contains the predictor names, “lower_bounds” and “upper_bounds” are the lower and upper bounds of beta, and “beta_given” is some supplied starting values for beta. | |
| Binary | A variable with only two possible outcomes. Refer to binomial. | |
| Binomial | A variable with the value 0 or 1. Binomial variables assigned as 0 indicate that an event hasn’t occurred or that the observation lacks a feature, where 1 indicates occurrence or instance of an attribute. | |
| Bins | Bins are linear-sized from the observed min-to-max for the subset that is being split. Large bins are enforced for shallow tree depths. Based on the tree decisions, as the tree gets deeper, the bins are distributed symmetrically over the reduced range of each subset. | |
| Categorical | A qualitative, unordered variable (for example, A, B, AB, and O would be values for the category blood type); synonym for enumerator or factor. Stored as an int column with a String mapping in H2O; limited to 10 million unique strings in H2O. |
|
| Classification | A model whose goal is to predict the category for the response input. | |
| Clip | In the H2O web UI Flow, a clip is a single cell in a flow containing an action that is saved for later reuse. | |
| Cloud | Synonym for cluster. Refer to the definition for cluster. | |
| Cluster | 1. A group of H2O nodes that work together; when a job is submitted to a cluster, all the nodes in the cluster work on a portion of the job. Synonym for cloud. 2. In statistics, a cluster is a group of observations from a data set identified as similar according to a particular clustering algorithm. |
|
| Confusion matrix | Table that depicts the performance of the algorithm (using the false positive rate, false negative, true positive, and true negative rates). | |
| Continuous | A variable that can take on all or nearly all values along an interval on the real number line (for example, height or weight). The opposite of a discrete value, which can only take on certain numerical values (for example, the number of patients treated). | |
| CSV file | CSV is an acronym for comma-separated value. A CSV file stores data in a plain text format. | |
| Deep Learning | Uses a composition of multiple non-linear transformations to model high-level abstractions in data. | |
| Dependent variable | The response column in H2O; what you are trying to measure, observe, or predict. The opposite of an independent variable. | |
| Data frame | A distributed representation of a large dataset. | |
| Destination key | Automatically generated key for a model that allows recall of a specific model later in analysis. Users can specify a different destination key than the key generated by H2O. | |
| Deviance | Deviance is the difference between an expected value and an observed value. It plays a critical role in defining GLM models. For a more detailed discussion of deviance, please refer to the H2O Data Science documentation on GLM. | |
| Distributed key/value (DKV) | Distributed key/value store. Refer also to key/value store. | |
| Discrete | A variable that can only take on certain numerical values (for example, the number of patients treated). The opposite of a continuous variable. | |
| Enumerator/enum | A data type where the value is one of a defined set of named values known as “elements”, “members”, or “enumerators.” For example, cat, dog, & mouse are enumerators of the enumerated type animal. | |
| Epoch | A round or iteration of model training or testing. Refer also to iteration. | |
| Factor | A data type where the value is one of a defined set of categories. Refer to Enum and Categorical. | |
| Family | The distribution options available for predictive modeling in GLM. | |
| Feature | Synonym for attribute, predictor, or independent variable. Usually refers to the data observed on features given in the columns of a data set. | |
| Feed-forward | Associates input with output for pattern recognition. | |
| Flatfile | A basic text file containing multiple IP addresses (one per line) used by H2O to configure a cluster. | |
| Flow | Refers to the series of cell-based actions created in H2O’s web UI or the web UI itself. | |
| Gzipped (gz) file | Gzip is a type of file compression commonly used for H2O file dependencies. | |
| HEX format | Records made up of hexadecimal numbers representing machine language code or constant data. In H2O, data must be parsed into .hex format before you can perform operations on it. | |
| Independent variable | The factors can be manipulated or controlled (also known as predictors). The opposite of a dependent variable. | |
| Hit ratio | (Multinomial only) The number of times the prediction was correct out of the total number of predictions. | |
| Instance | Occurs each time H2O is started. This process builds a cluster of nodes (even if it is only a one-node cluster on a local machine). The instance begins when the cluster is formed and ends when the program is closed. | |
| Integer | A whole number (can be negative but cannot be a fraction). Can be represented in H2O as an int, which is not a type but a property of the data. |
|
| Iteration | A round or instance of model testing or training. Also known as an epoch. | |
| Job | A task performed by H2O. For example, reading a data file, parsing a data file, or building a model. In the browser-based GUI of H2O, each job is listed in the Admin menu under Jobs. | |
| JVM | Java virtual machine; used to run H2O. | |
| Key | The .hex key generated when data are parsed into H2O. In the web-based GUI, key is an input on each page where users define models and any page where users validate models on a new data set or use a model to generate predictions. | |
| Key/value pair | A type of data that associates a particular key index to a certain datum. | |
| Key/value store | A tool that allows storage of schema-less data. Data usually consists of a string that represents the key, and the data itself, which is the value. Refer also to distributed key/value. | |
| L1 regularization | A regularization method that constrains the absolute value of the weights and has the net effect of dropping some values (setting them to zero) from a model to reduce complexity and avoid overfitting. | |
| L2 regularization | A regularization method that constrains the sum of the squared weights. This method introduces bias into parameter estimates but frequently produces substantial gains in modeling as estimate variance is reduced. | |
| Link function | A user-defined option in GLM. | |
| Loss function | The function minimized in order to achieve a desired estimator; synonymous to objective function and criterion function. For example, linear regression defines the set of best parameter estimates as the set of estimates that produces the minimum of the sum of the squared errors. Errors are the difference between the predicted value and the observed value. | |
| MSE | Mean squared error; measures the average of the squares of the error rate (the difference between the predictors and what was predicted). | |
| Multinomial | A variable where the value can be one of more than two possible outcomes (for example, blood type). | |
| N-folds | User-defined number of cross validation models generated by H2O. | |
| Node | In distributed computing systems, nodes include clients,servers, or peers. In statistics, a node is a decision or terminal point in a classification tree. | |
| Numeric | A column type containing real numbers, small integers, or booleans. | |
| Offset | A parameter that compensates for differences in units of observation (for example, different populations or geographic sizes) to make sure outcome is proportional. | |
| Outline | In H2O’s web UI Flow, a brief summary of the actions contained in the cells. | |
| Parse | Analysis of a string of symbols or datum that results in the conversion of a set of information from a person-readable format to a machine-readable format. | |
| POJO | Plain Old Java Object; a way to export a model built in H2O and implement it in a Java application. | |
| Regression | A model where the input is numerical and the output is a prediction of numerical values. Also known as “quantitative”; the opposite of a classification model. | |
| Response column | Method of selecting the dependent variable in H2O. | |
| Real | A fractional number. | |
| ROC Curve | Graph representing the ratio to true positives to false positives. | |
| Scoring history | Represents the error rate of the model as it is built. | |
| Seed | A starting point for randomization. Seed specification is used when machine learning models have a random component; it allows users to recreate the exact “random” conditions used in a model at a later time. | |
| Separator | What separates the entries in the dataset; usually a comma, semicolon, etc. | |
| Sparse | A dataset where many of the rows contain blank values or “NA” instead of data. | |
| Standard deviation | The standard deviation of the data in the column, defined as the square root of the sum of the deviance of observed values from the mean divided by the number of elements in the column minus one. Abbreviated sd. | |
| Standardization | Transformation of a variable so that it is mean-centered at 0 and scaled by the standard deviation; helps prevent precision problems. | |
| String | Refers to data where each entry is typically unique (for example, a dataset containing people’s names and addresses). | |
| Supervised learning | Model type where the input is labeled so that the algorithm can identify it and learn from it. | |
| Time | Data type supported by H2O; represented as “milliseconds-since-the-Unix-Epoch”; stored internally as a 64-bit integer in a standard int column. Used directly by the Cox Proportional Hazards model but also used to build other features. |
|
| Training frame | The dataset used to build the model. | |
| Unsupervised learning | Model type where the input is not labeled. | |
| UUID | A dense representation of universally unique identifiers (UUIDs) used to label and group events; stored as a 128-bit numeric value. | |
| Validation | An analysis of how well the model fits. | |
| Validation frame | The dataset used to evaluate the accuracy of the model. | |
| Variable importance | Represents the statistical significance of each variable in the data in terms of its affect on the model. | |
| Weights | A parameter that specifies certain outcomes as more significant (for example, if you are trying to identify incidence of disease, one “positive” result can be more meaningful than 50 “negative” responses). Higher values indicate more importance. | |
| XLS file | A Microsoft Excel 2003-2007 spreadsheet file format. | |
| Y | Dependent variable used in GLM; a user-defined input selected from the set of variables present in the user’s data. | |
| YARN | Yet Another Resource Manager; used to manage H2O on a Hadoop cluster. |
Quick Start Videos
- H2O Quick Start with Flow
- H2O Quick Start with Python
- H2O Quick Start on Hadoop
- H2O Quick Start with Sparkling Water
- H2O Quick Start with R
H2O Quick Start with Flow
H2O Quick Start with Python
H2O Quick Start on Hadoop
H2O Quick Start with Sparkling Water
H2O Quick Start with R
REST API Reference
- /3/About
- /3/Cloud
- /3/Cloud
- /3/CreateFrame
- /3/DKV
- /3/DKV/(?
.*) - /3/DataInfoFrame
- /3/DownloadDataset
- /3/DownloadDataset.bin
- /3/Find
- /3/Frames
- /3/Frames
- /3/Frames/(?
.*) - /3/Frames/(?
.*) - /3/Frames/(?
.*)/columns - /3/Frames/(?
.*)/columns/(? .*) - /3/Frames/(?
.*)/columns/(? .*)/domain - /3/Frames/(?
.*)/columns/(? .*)/summary - /3/Frames/(?
.*)/export - /3/Frames/(?
.*)/export/(? .*)/overwrite/(? .*) - /3/Frames/(?
.*)/summary - /3/GarbageCollect
- /3/ImportFiles
- /3/ImportFiles
- /3/InitID
- /3/InitID
- /3/Interaction
- /3/JStack
- /3/Jobs
- /3/Jobs/(?
.*) - /3/Jobs/(?
.*)/cancel - /3/KillMinus3
- /3/LogAndEcho
- /3/Logs/nodes/(?
.*)/files/(? .*) - /3/MakeGLMModel
- /3/Metadata/endpoints
- /3/Metadata/endpoints/(?
[0-9]+) - /3/Metadata/endpoints/(?
.*) - /3/Metadata/schemaclasses/(?
.*) - /3/Metadata/schemas
- /3/Metadata/schemas/(?
.*) - /3/MissingInserter
- /3/ModelBuilders
- /3/ModelBuilders/(?
.*) - /3/ModelBuilders/(?
.*)/model_id - /3/ModelBuilders/aggregator
- /3/ModelBuilders/aggregator/parameters
- /3/ModelBuilders/deeplearning
- /3/ModelBuilders/deeplearning/parameters
- /3/ModelBuilders/drf
- /3/ModelBuilders/drf/parameters
- /3/ModelBuilders/gbm
- /3/ModelBuilders/gbm/parameters
- /3/ModelBuilders/glm
- /3/ModelBuilders/glm/parameters
- /3/ModelBuilders/glrm
- /3/ModelBuilders/glrm/parameters
- /3/ModelBuilders/kmeans
- /3/ModelBuilders/kmeans/parameters
- /3/ModelBuilders/naivebayes
- /3/ModelBuilders/naivebayes/parameters
- /3/ModelBuilders/pca
- /3/ModelBuilders/pca/parameters
- /3/ModelMetrics
- /3/ModelMetrics/frames/(?.*)
- /3/ModelMetrics/frames/(?.*)/models/(?
.*) - /3/ModelMetrics/frames/(?.*)/models/(?
.*) - /3/ModelMetrics/models/(?
.*) - /3/ModelMetrics/models/(?
.*)/frames/(?.*) - /3/ModelMetrics/models/(?
.*)/frames/(?.*) - /3/ModelMetrics/models/(?
.*)/frames/(?.*) - /3/Models
- /3/Models
- /3/Models.java/(?
.*) - /3/Models.java/(?
.*)/preview - /3/Models/(?
.*) - /3/Models/(?
.*) - /3/NetworkTest
- /3/NodePersistentStorage/(?
.*) - /3/NodePersistentStorage/(?
.*) - /3/NodePersistentStorage/(?
.*)/(? .*) - /3/NodePersistentStorage/(?
.*)/(? .*) - /3/NodePersistentStorage/(?
.*)/(? .*) - /3/NodePersistentStorage/categories/(?
.*)/exists - /3/NodePersistentStorage/categories/(?
.*)/names/(? .*)/exists - /3/NodePersistentStorage/configured
- /3/Parse
- /3/ParseSVMLight
- /3/ParseSetup
- /3/Predictions/models/(?
.*)/frames/(?.*) - /3/Profiler
- /3/Shutdown
- /3/SplitFrame
- /3/Timeline
- /3/Typeahead/files
- /3/UnlockKeys
- /3/WaterMeterCpuTicks/(?
.*) - /3/WaterMeterIo
- /3/WaterMeterIo/(?
.*) - /4/Predictions/models/(?
.*)/frames/(?.*) - /99/Assembly
- /99/Assembly.java/(?
.*)/(? .*) - /99/DCTTransformer
- /99/Grid/aggregator
- /99/Grid/deeplearning
- /99/Grid/drf
- /99/Grid/gbm
- /99/Grid/glm
- /99/Grid/glrm
- /99/Grid/kmeans
- /99/Grid/naivebayes
- /99/Grid/pca
- /99/Grid/svd
- /99/Grids
- /99/Grids/(?
.*) - /99/ImportSQLTable
- /99/ModelBuilders/svd
- /99/ModelBuilders/svd/parameters
- /99/Models.bin/(?
.*) - /99/Models.bin/(?
.*) - /99/Rapids
- /99/Sample
- /99/Tabulate
GET /3/About
Return information about this H2O cluster.
| Input | AboutV3 |
| Output | AboutV3 |
GET /3/Cloud
Determine the status of the nodes in the H2O cloud.
| Input | CloudV3 |
| Output | CloudV3 |
HEAD /3/Cloud
Determine the status of the nodes in the H2O cloud.
| Input | CloudV3 |
| Output | CloudV3 |
POST /3/CreateFrame
Create a synthetic H2O Frame.
| Input | CreateFrameV3 |
| Output | JobV3 |
DELETE /3/DKV
Remove all keys from the H2O distributed K/V store.
| Input | RemoveAllV3 |
| Output | RemoveAllV3 |
DELETE /3/DKV/(?.*)
Remove an arbitrary key from the H2O distributed K/V store.
| Input | RemoveV3 |
| Output | RemoveV3 |
POST /3/DataInfoFrame
test only
| Input | DataInfoFrameV3 |
| Output | DataInfoFrameV3 |
GET /3/DownloadDataset
Download dataset as a CSV.
| Input | DownloadDataV3 |
| Output | DownloadDataV3 |
GET /3/DownloadDataset.bin
Download dataset as a CSV.
| Input | DownloadDataV3 |
| Output | DownloadDataV3 |
GET /3/Find
Find a value within a Frame.
| Input | FindV3 |
| Output | FindV3 |
GET /3/Frames
Return all Frames in the H2O distributed K/V store.
| Input | FramesV3 |
| Output | FramesV3 |
DELETE /3/Frames
Delete all Frames from the H2O distributed K/V store.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)
Return the specified Frame.
| Input | FramesV3 |
| Output | FramesV3 |
DELETE /3/Frames/(?.*)
Delete the specified Frame from the H2O distributed K/V store.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)/columns
Return all the columns from a Frame.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)/columns/(?.*)
Return the specified column from a Frame.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)/columns/(?.*)/domain
Return the domains for the specified column. “null” if the column is not a categorical.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)/columns/(?.*)/summary
Return the summary metrics for a column, e.g. mins, maxes, mean, sigma, percentiles, etc.
| Input | FramesV3 |
| Output | FramesV3 |
POST /3/Frames/(?.*)/export
Export a Frame to the given path with optional overwrite.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)/export/(?.*)/overwrite/(?.*)
Export a Frame to the given path with optional overwrite.
| Input | FramesV3 |
| Output | FramesV3 |
GET /3/Frames/(?.*)/summary
Return a Frame, including the histograms, after forcing computation of rollups.
| Input | FramesV3 |
| Output | FramesV3 |
POST /3/GarbageCollect
Explicitly call System.gc().
| Input | GarbageCollectV3 |
| Output | GarbageCollectV3 |
GET /3/ImportFiles
Import raw data files into a single-column H2O Frame.
| Input | ImportFilesV3 |
| Output | ImportFilesV3 |
POST /3/ImportFiles
Import raw data files into a single-column H2O Frame.
| Input | ImportFilesV3 |
| Output | ImportFilesV3 |
GET /3/InitID
Issue a new session ID.
| Input | InitIDV3 |
| Output | InitIDV3 |
DELETE /3/InitID
End a session.
| Input | InitIDV3 |
| Output | InitIDV3 |
POST /3/Interaction
Create interactions between categorical columns.
| Input | InteractionV3 |
| Output | JobV3 |
GET /3/JStack
Report stack traces for all threads on all nodes.
| Input | JStackV3 |
| Output | JStackV3 |
GET /3/Jobs
Get a list of all the H2O Jobs (long-running actions).
| Input | JobsV3 |
| Output | JobsV3 |
GET /3/Jobs/(?.*)
Get the status of the given H2O Job (long-running action).
| Input | JobsV3 |
| Output | JobsV3 |
POST /3/Jobs/(?.*)/cancel
Cancel a running job.
| Input | JobsV3 |
| Output | JobsV3 |
GET /3/KillMinus3
Kill minus 3 on this node
| Input | KillMinus3V3 |
| Output | KillMinus3V3 |
POST /3/LogAndEcho
Save a message to the H2O logfile.
| Input | LogAndEchoV3 |
| Output | LogAndEchoV3 |
GET /3/Logs/nodes/(?.*)/files/(?.*)
Get named log file for a node.
| Input | LogsV3 |
| Output | LogsV3 |
POST /3/MakeGLMModel
make a new GLM model based on existing one
| Input | MakeGLMModelV3 |
| Output | GLMModelV3 |
GET /3/Metadata/endpoints
Return a list of all the REST API endpoints.
| Input | MetadataV3 |
| Output | MetadataV3 |
GET /3/Metadata/endpoints/(?[0-9]+)
Return the REST API endpoint metadata, including documentation, for the endpoint specified by number.
| Input | MetadataV3 |
| Output | MetadataV3 |
GET /3/Metadata/endpoints/(?.*)
Return the REST API endpoint metadata, including documentation, for the endpoint specified by path.
| Input | MetadataV3 |
| Output | MetadataV3 |
GET /3/Metadata/schemaclasses/(?.*)
Return the REST API schema metadata for specified schema class.
| Input | MetadataV3 |
| Output | MetadataV3 |
GET /3/Metadata/schemas
Return list of all REST API schemas.
| Input | MetadataV3 |
| Output | MetadataV3 |
GET /3/Metadata/schemas/(?.*)
Return the REST API schema metadata for specified schema.
| Input | MetadataV3 |
| Output | MetadataV3 |
POST /3/MissingInserter
Insert missing values.
| Input | MissingInserterV3 |
| Output | JobV3 |
GET /3/ModelBuilders
Return the Model Builder metadata for all available algorithms.
| Input | ModelBuildersV3 |
| Output | ModelBuildersV3 |
GET /3/ModelBuilders/(?.*)
Return the Model Builder metadata for the specified algorithm.
| Input | ModelBuildersV3 |
| Output | ModelBuildersV3 |
POST /3/ModelBuilders/(?.*)/model_id
Return a new unique model_id for the specified algorithm.
| Input | ModelBuildersV3 |
| Output | ModelIdV3 |
POST /3/ModelBuilders/aggregator
Train a Aggregator model.
| Input | AggregatorV3 |
| Output | AggregatorV3 |
POST /3/ModelBuilders/aggregator/parameters
Validate a set of Aggregator model builder parameters.
| Input | AggregatorV3 |
| Output | AggregatorV3 |
POST /3/ModelBuilders/deeplearning
Train a DeepLearning model.
| Input | DeepLearningV3 |
| Output | DeepLearningV3 |
POST /3/ModelBuilders/deeplearning/parameters
Validate a set of DeepLearning model builder parameters.
| Input | DeepLearningV3 |
| Output | DeepLearningV3 |
POST /3/ModelBuilders/drf
Train a DRF model.
| Input | DRFV3 |
| Output | DRFV3 |
POST /3/ModelBuilders/drf/parameters
Validate a set of DRF model builder parameters.
| Input | DRFV3 |
| Output | DRFV3 |
POST /3/ModelBuilders/gbm
Train a GBM model.
| Input | GBMV3 |
| Output | GBMV3 |
POST /3/ModelBuilders/gbm/parameters
Validate a set of GBM model builder parameters.
| Input | GBMV3 |
| Output | GBMV3 |
POST /3/ModelBuilders/glm
Train a GLM model.
| Input | GLMV3 |
| Output | GLMV3 |
POST /3/ModelBuilders/glm/parameters
Validate a set of GLM model builder parameters.
| Input | GLMV3 |
| Output | GLMV3 |
POST /3/ModelBuilders/glrm
Train a GLRM model.
| Input | GLRMV3 |
| Output | GLRMV3 |
POST /3/ModelBuilders/glrm/parameters
Validate a set of GLRM model builder parameters.
| Input | GLRMV3 |
| Output | GLRMV3 |
POST /3/ModelBuilders/kmeans
Train a KMeans model.
| Input | KMeansV3 |
| Output | KMeansV3 |
POST /3/ModelBuilders/kmeans/parameters
Validate a set of KMeans model builder parameters.
| Input | KMeansV3 |
| Output | KMeansV3 |
POST /3/ModelBuilders/naivebayes
Train a NaiveBayes model.
| Input | NaiveBayesV3 |
| Output | NaiveBayesV3 |
POST /3/ModelBuilders/naivebayes/parameters
Validate a set of NaiveBayes model builder parameters.
| Input | NaiveBayesV3 |
| Output | NaiveBayesV3 |
POST /3/ModelBuilders/pca
Train a PCA model.
| Input | PCAV3 |
| Output | PCAV3 |
POST /3/ModelBuilders/pca/parameters
Validate a set of PCA model builder parameters.
| Input | PCAV3 |
| Output | PCAV3 |
GET /3/ModelMetrics
Return all the saved scoring metrics.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
GET /3/ModelMetrics/frames/(?.*)
Return the saved scoring metrics for the specified Frame.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
GET /3/ModelMetrics/frames/(?.*)/models/(?.*)
Return the saved scoring metrics for the specified Model and Frame.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
DELETE /3/ModelMetrics/frames/(?.*)/models/(?.*)
Return the saved scoring metrics for the specified Model and Frame.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
GET /3/ModelMetrics/models/(?.*)
Return the saved scoring metrics for the specified Model.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
GET /3/ModelMetrics/models/(?.*)/frames/(?.*)
Return the saved scoring metrics for the specified Model and Frame.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
DELETE /3/ModelMetrics/models/(?.*)/frames/(?.*)
Return the saved scoring metrics for the specified Model and Frame.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
POST /3/ModelMetrics/models/(?.*)/frames/(?.*)
Return the scoring metrics for the specified Frame with the specified Model. If the Frame has already been scored with the Model then cached results will be returned; otherwise predictions for all rows in the Frame will be generated and the metrics will be returned.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
GET /3/Models
Return all Models from the H2O distributed K/V store.
| Input | ModelsV3 |
| Output | ModelsV3 |
DELETE /3/Models
Delete all Models from the H2O distributed K/V store.
| Input | ModelsV3 |
| Output | ModelsV3 |
GET /3/Models.java/(?.*)
Return the stream containing model implementation in Java code.
| Input | ModelsV3 |
| Output | StreamingSchema |
GET /3/Models.java/(?.*)/preview
Return potentially abridged model suitable for viewing in a browser (currently only used for java model code).
| Input | ModelsV3 |
| Output | StreamingSchema |
GET /3/Models/(?.*)
Return the specified Model from the H2O distributed K/V store, optionally with the list of compatible Frames.
| Input | ModelsV3 |
| Output | ModelsV3 |
DELETE /3/Models/(?.*)
Delete the specified Model from the H2O distributed K/V store.
| Input | ModelsV3 |
| Output | ModelsV3 |
GET /3/NetworkTest
Run a network test to measure the performance of the cluster interconnect.
| Input | NetworkTestV3 |
| Output | NetworkTestV3 |
POST /3/NodePersistentStorage/(?.*)
Store a value.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
GET /3/NodePersistentStorage/(?.*)
Return all keys stored for a given category.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
POST /3/NodePersistentStorage/(?.*)/(?.*)
Store a named value.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
GET /3/NodePersistentStorage/(?.*)/(?.*)
Return value for a given name.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
DELETE /3/NodePersistentStorage/(?.*)/(?.*)
Delete a key.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
GET /3/NodePersistentStorage/categories/(?.*)/exists
Return true or false.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
GET /3/NodePersistentStorage/categories/(?.*)/names/(?.*)/exists
Return true or false.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
GET /3/NodePersistentStorage/configured
Return true or false.
| Input | NodePersistentStorageV3 |
| Output | NodePersistentStorageV3 |
POST /3/Parse
Parse a raw byte-oriented Frame into a useful columnar data Frame.
| Input | ParseV3 |
| Output | ParseV3 |
POST /3/ParseSVMLight
Parse a raw byte-oriented Frame into a useful columnar data Frame.
| Input | ParseSVMLightV3 |
| Output | JobV3 |
POST /3/ParseSetup
Guess the parameters for parsing raw byte-oriented data into an H2O Frame.
| Input | ParseSetupV3 |
| Output | ParseSetupV3 |
POST /3/Predictions/models/(?.*)/frames/(?.*)
Score (generate predictions) for the specified Frame with the specified Model. Both the Frame of predictions and the metrics will be returned.
| Input | ModelMetricsListSchemaV3 |
| Output | ModelMetricsListSchemaV3 |
GET /3/Profiler
Report real-time profiling information for all nodes (sorted, aggregated stack traces).
| Input | ProfilerV3 |
| Output | ProfilerV3 |
POST /3/Shutdown
Shut down the cluster
| Input | ShutdownV3 |
| Output | ShutdownV3 |
POST /3/SplitFrame
Split a H2O Frame.
| Input | SplitFrameV3 |
| Output | SplitFrameV3 |
GET /3/Timeline
Debugging tool that provides information on current communication between nodes.
| Input | TimelineV3 |
| Output | TimelineV3 |
GET /3/Typeahead/files
Typehead hander for filename completion.
| Input | TypeaheadV3 |
| Output | TypeaheadV3 |
POST /3/UnlockKeys
Unlock all keys in the H2O distributed K/V store, to attempt to recover from a crash.
| Input | UnlockKeysV3 |
| Output | UnlockKeysV3 |
GET /3/WaterMeterCpuTicks/(?.*)
Return a CPU usage snapshot of all cores of all nodes in the H2O cluster.
| Input | WaterMeterCpuTicksV3 |
| Output | WaterMeterCpuTicksV3 |
GET /3/WaterMeterIo
Return IO usage snapshot of all nodes in the H2O cluster.
| Input | WaterMeterIoV3 |
| Output | WaterMeterIoV3 |
GET /3/WaterMeterIo/(?.*)
Return IO usage snapshot of all nodes in the H2O cluster.
| Input | WaterMeterIoV3 |
| Output | WaterMeterIoV3 |
POST /4/Predictions/models/(?.*)/frames/(?.*)
Score (generate predictions) for the specified Frame with the specified Model. Both the Frame of predictions and the metrics will be returned.
| Input | ModelMetricsListSchemaV3 |
| Output | JobV3 |
POST /99/Assembly
Fit an assembly to an input frame
| Input | AssemblyV99 |
| Output | AssemblyV99 |
GET /99/Assembly.java/(?.*)/(?.*)
Generate a Java POJO from the Assembly
| Input | AssemblyV99 |
| Output | AssemblyV99 |
POST /99/DCTTransformer
Row-by-Row discrete cosine transforms in 1D, 2D and 3D.
| Input | DCTTransformerV3 |
| Output | JobV3 |
POST /99/Grid/aggregator
Run grid search for Aggregator model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/deeplearning
Run grid search for DeepLearning model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/drf
Run grid search for DRF model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/gbm
Run grid search for GBM model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/glm
Run grid search for GLM model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/glrm
Run grid search for GLRM model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/kmeans
Run grid search for KMeans model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/naivebayes
Run grid search for NaiveBayes model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/pca
Run grid search for PCA model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
POST /99/Grid/svd
Run grid search for SVD model.
| Input | GridSearchSchema |
| Output | GridSearchSchema |
GET /99/Grids
Return all grids from H2O distributed K/V store.
| Input | GridsV99 |
| Output | GridsV99 |
GET /99/Grids/(?.*)
Return the specified grid search result.
| Input | GridSchemaV99 |
| Output | GridSchemaV99 |
POST /99/ImportSQLTable
Import SQL table into an H2O Frame.
| Input | ImportSQLTableV99 |
| Output | JobV3 |
POST /99/ModelBuilders/svd
Train a SVD model.
| Input | SVDV99 |
| Output | SVDV99 |
POST /99/ModelBuilders/svd/parameters
Validate a set of SVD model builder parameters.
| Input | SVDV99 |
| Output | SVDV99 |
POST /99/Models.bin/(?.*)
Import given binary model into H2O.
| Input | ModelImportV3 |
| Output | ModelsV3 |
GET /99/Models.bin/(?.*)
Export given model.
| Input | ModelExportV3 |
| Output | ModelExportV3 |
POST /99/Rapids
Execute an Rapids AST.
| Input | RapidsSchema |
| Output | RapidsSchema |
GET /99/Sample
Example of an experimental endpoint. Call via /EXPERIMENTAL/Sample. Experimental endpoints can change at any moment.
| Input | CloudV3 |
| Output | CloudV3 |
POST /99/Tabulate
Tabulate one column vs another.
| Input | TabulateV3 |
| Output | TabulateV3 |
REST API Schema Reference
- AboutEntryV3
- AboutV3
- AggregatorModelOutputV3
- AggregatorModelV3
- AggregatorParametersV3
- AggregatorV3
- AssemblyKeyV3
- AssemblyV99
- CartesianSearchCriteriaV99
- CloudV3
- ClusteringModelBuilderSchema
- ClusteringModelParametersSchema
- ColSpecifierV3
- ColV3
- ColumnSpecsBase
- ConfusionMatrixBase
- ConfusionMatrixV3
- CoxPHModelOutputV3
- CoxPHModelV3
- CoxPHParametersV3
- CoxPHV3
- CreateFrameV3
- DCTTransformerV3
- DRFModelOutputV3
- DRFModelV3
- DRFParametersV3
- DRFV3
- DStackTraceV3
- DataInfoFrameV3
- DeepLearningModelOutputV3
- DeepLearningModelV3
- DeepLearningParametersV3
- DeepLearningV3
- DownloadDataV3
- EventV3
- ExampleModelOutputV3
- ExampleModelV3
- ExampleParametersV3
- ExampleV3
- FieldMetadataBase
- FieldMetadataV3
- FindV3
- FrameBase
- FrameKeyV3
- FrameSynopsisV3
- FrameV3
- FramesBase
- FramesV3
- GBMModelOutputV3
- GBMModelV3
- GBMParametersV3
- GBMV3
- GLMModelOutputV3
- GLMModelV3
- GLMParametersV3
- GLMV3
- GLRMModelOutputV3
- GLRMModelV3
- GLRMParametersV3
- GLRMV3
- GarbageCollectV3
- GrepModelOutputV3
- GrepModelV3
- GrepParametersV3
- GrepV3
- GridKeyV3
- GridSchemaV99
- GridSearchSchema
- GridsV99
- H2OErrorV3
- H2OModelBuilderErrorV3
- HeartBeatEvent
- HyperSpaceSearchCriteriaV99
- IOEvent
- ImportFilesV3
- ImportSQLTableV99
- InitIDV3
- InteractionV3
- IoStatsEntry
- JStackV3
- JobKeyV3
- JobV3
- JobsV3
- KMeansModelOutputV3
- KMeansModelV3
- KMeansParametersV3
- KMeansV3
- KeyV3
- KillMinus3V3
- LogAndEchoV3
- LogsV3
- MakeGLMModelV3
- MetadataBase
- MetadataV3
- MissingInserterV3
- ModelBuilderSchema
- ModelBuilderV3
- ModelBuildersBase
- ModelBuildersV3
- ModelExportV3
- ModelIdV3
- ModelImportV3
- ModelKeyV3
- ModelMetricsAutoEncoderV3
- ModelMetricsBase
- ModelMetricsBinomialGLMV3
- ModelMetricsBinomialV3
- ModelMetricsClusteringV3
- ModelMetricsGLRMV99
- ModelMetricsListSchemaV3
- ModelMetricsMultinomialGLMV3
- ModelMetricsMultinomialV3
- ModelMetricsPCAV3
- ModelMetricsRegressionGLMV3
- ModelMetricsRegressionV3
- ModelMetricsSVDV99
- ModelOutputSchema
- ModelParameterSchemaV3
- ModelParametersSchema
- ModelSchema
- ModelSchemaBase
- ModelSynopsisV3
- ModelsBase
- ModelsV3
- NaiveBayesModelOutputV3
- NaiveBayesModelV3
- NaiveBayesParametersV3
- NaiveBayesV3
- NetworkBenchV3
- NetworkEvent
- NetworkTestV3
- NodePersistentStorageEntryV3
- NodePersistentStorageV3
- NodeV3
- PCAModelOutputV3
- PCAModelV3
- PCAParametersV3
- PCAV3
- ParseSVMLightV3
- ParseSetupV3
- ParseV3
- ProfilerNodeEntryV3
- ProfilerNodeV3
- ProfilerV3
- QuantileParametersV3
- QuantileV3
- RandomDiscreteValueSearchCriteriaV99
- RapidsFrameV3
- RapidsFunctionV3
- RapidsNumberV3
- RapidsNumbersV3
- RapidsSchema
- RapidsStringV3
- RapidsStringsV3
- RapidsV99
- RemoveAllV3
- RemoveV3
- RequestSchema
- RouteBase
- RouteV3
- SVDModelOutputV99
- SVDModelV99
- SVDParametersV99
- SVDV99
- Schema
- SchemaMetadataBase
- SchemaMetadataV3
- SharedTreeModelOutputV3
- SharedTreeModelV3
- SharedTreeParametersV3
- SharedTreeV3
- ShutdownV3
- SplitFrameV3
- StreamingSchema
- SynonymV3
- TabulateV3
- TimelineV3
- TreeStatsV3
- TwoDimTableBase
- TwoDimTableV3
- TypeaheadV3
- UnlockKeysV3
- ValidationMessageBase
- ValidationMessageV3
- VarImpBase
- VarImpV3
- WaterMeterCpuTicksV3
- WaterMeterIoV3
- Word2VecModelOutputV3
- Word2VecModelV3
- Word2VecParametersV3
- Word2VecV3
AboutEntryV3
namestring | Property name | Out |
valuestring | Property value | Out |
AboutV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
entriesIced[] | List of properties about this running H2O instance | Out |
AggregatorModelOutputV3
output_frameKey | Aggregated Frame of Exemplars | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
AggregatorModelV3
model_idKey | Model key | In/Out |
parametersAggregatorParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputAggregatorOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
AggregatorParametersV3
radius_scaledouble | Radius scaling | In |
transformenum | Transformation of training data | In |
pca_methodenum | Method for computing PCA (Caution: Power and GLRM are currently experimental and unstable) | In |
kint | Rank of matrix approximation | In/Out |
max_iterationsint | Maximum number of iterations for PCA | In/Out |
seedlong | RNG seed for initialization | In/Out |
use_all_factor_levelsboolean | Whether first factor level is included in each categorical expansion | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
AggregatorV3
parametersAggregatorParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
AssemblyKeyV3
namestring | Name (string representation) for this Key. | In/Out |
typestring | Name (string representation) for the type of Keyed this Key points to. | In/Out |
URLstring | URL for the resource that this Key points to, if one exists. | In/Out |
AssemblyV99
stepsstring[] | A list of steps describing the assembly line. | In |
frameKey | Input Frame for the assembly. | In |
pojo_namestring | The name of the file and generated class | In |
assembly_idstring | The key of the Assembly object to retrieve from the DKV. | In |
resultKey | Output of the assembly line. | In |
assemblyKey | A Key to the fit Assembly data structure | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
CartesianSearchCriteriaV99
strategyenum | Hyperparameter space search strategy. | In/Out |
CloudV3
skip_ticksboolean | skip_ticks | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
versionstring | version | Out |
branch_namestring | branch_name | Out |
build_numberstring | build_number | Out |
node_idxint | Node index number cloud status is collected from (zero-based) | Out |
cloud_namestring | cloud_name | Out |
cloud_sizeint | cloud_size | Out |
cloud_uptime_millislong | cloud_uptime_millis | Out |
cloud_healthyboolean | cloud_healthy | Out |
bad_nodesint | Nodes reporting unhealthy | Out |
consensusboolean | Cloud voting is stable | Out |
lockedboolean | Cloud is accepting new members or not | Out |
is_clientboolean | Cloud is in client mode. | Out |
nodesIced[] | nodes | Out |
ClusteringModelBuilderSchema
parametersParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
ClusteringModelParametersSchema
kint | Number of clusters | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
ColSpecifierV3
column_namestring | Name of the column | In/Out |
is_member_of_framesstring[] | List of fields which specify columns that must contain this column | In/Out |
ColV3
labelstring | label | Out |
missing_countlong | missing | Out |
zero_countlong | zeros | Out |
positive_infinity_countlong | positive infinities | Out |
negative_infinity_countlong | negative infinities | Out |
minsdouble[] | mins | Out |
maxsdouble[] | maxs | Out |
meandouble | mean | Out |
sigmadouble | sigma | Out |
typestring | datatype: {enum, string, int, real, time, uuid} | Out |
domainstring[] | domain; not-null for categorical columns only | Out |
domain_cardinalityint | cardinality of this column’s domain; not-null for categorical columns only | Out |
datadouble[] | data | Out |
string_datastring[] | string data | Out |
precisionbyte | decimal precision, -1 for all digits | Out |
histogram_binslong[] | Histogram bins; null if not computed | Out |
histogram_basedouble | Start of histogram bin zero | Out |
histogram_stridedouble | Stride per bin | Out |
percentilesdouble[] | Percentile values, matching the default percentiles | Out |
ColumnSpecsBase
namestring | Column Name | Out |
typestring | Column Type | Out |
formatstring | Column Format (printf) | Out |
descriptionstring | Column Description | Out |
ConfusionMatrixBase
tableTwoDimTable | Annotated confusion matrix | Out |
ConfusionMatrixV3
tableTwoDimTable | Annotated confusion matrix | Out |
CoxPHModelOutputV3
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
CoxPHModelV3
model_idKey | Model key | In/Out |
parametersCoxPHParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputCoxPHOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
CoxPHParametersV3
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
CoxPHV3
parametersCoxPHParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
CreateFrameV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
destKey | destination key | In/Out |
rowslong | Number of rows | In/Out |
colsint | Number of data columns (in addition to the first response column) | In/Out |
seedlong | Random number seed that determines the random values | In/Out |
seed_for_column_typeslong | Random number seed for setting the column types | In/Out |
randomizeboolean | Whether frame should be randomized | In/Out |
valuelong | Constant value (for randomize=false) | In/Out |
real_rangelong | Range for real variables (-range … range) | In/Out |
categorical_fractiondouble | Fraction of categorical columns (for randomize=true) | In/Out |
factorsint | Factor levels for categorical variables | In/Out |
integer_fractiondouble | Fraction of integer columns (for randomize=true) | In/Out |
integer_rangelong | Range for integer variables (-range … range) | In/Out |
binary_fractiondouble | Fraction of binary columns (for randomize=true) | In/Out |
binary_ones_fractiondouble | Fraction of 1’s in binary columns | In/Out |
time_fractiondouble | Fraction of date/time columns (for randomize=true) | In/Out |
string_fractiondouble | Fraction of string columns (for randomize=true) | In/Out |
missing_fractiondouble | Fraction of missing values | In/Out |
response_factorsint | Number of factor levels of the first column (1=real, 2=binomial, N=multinomial) | In/Out |
has_responseboolean | Whether an additional response column should be generated | In/Out |
keyKey | Job Key | Out |
DCTTransformerV3
datasetKey | Dataset | In |
destination_frameKey | Destination Frame ID | In |
dimensionsint[] | Dimensions of the input array: Height, Width, Depth (Nx1x1 for 1D, NxMx1 for 2D) | In |
inverseboolean | Whether to do the inverse transform | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
DRFModelOutputV3
variable_importancesTwoDimTable | Variable Importances | Out |
init_fdouble | The Intercept term, the initial model function value to which trees make adjustments | Out |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
DRFModelV3
model_idKey | Model key | In/Out |
parametersDRFParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputDRFOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
DRFParametersV3
mtriesint | Number of variables randomly sampled as candidates at each split. If set to -1, defaults to sqrt{p} for classification and p/3 for regression (where p is the # of predictors | In |
binomial_double_treesboolean | For binary classification: Build 2x as many trees (one per class) - can lead to higher accuracy. | In |
ntreesint | Number of trees. | In |
max_depthint | Maximum tree depth. | In |
min_rowsdouble | Fewest allowed (weighted) observations in a leaf (in R called ‘nodesize’). | In |
nbinsint | For numerical columns (real/int), build a histogram of (at least) this many bins, then split at the best point | In |
nbins_top_levelint | For numerical columns (real/int), build a histogram of (at most) this many bins at the root level, then decrease by factor of two per level | In |
nbins_catsint | For categorical columns (factors), build a histogram of this many bins, then split at the best point. Higher values can lead to more overfitting. | In |
r2_stoppingdouble | Stop making trees when the R^2 metric equals or exceeds this | In |
seedlong | Seed for pseudo random number generator (if applicable) | In |
build_tree_one_nodeboolean | Run on one node only; no network overhead but fewer cpus used. Suitable for small datasets. | In |
sample_ratedouble | Row sample rate per tree (from 0.0 to 1.0) | In |
sample_rate_per_classdouble[] | Row sample rate per tree per class (from 0.0 to 1.0) | In |
col_sample_rate_per_treedouble | Column sample rate per tree (from 0.0 to 1.0) | In |
col_sample_rate_change_per_leveldouble | Relative change of the column sampling rate for every level (from 0.0 to 2.0) | In |
score_tree_intervalint | Score the model after every so many trees. Disabled if set to 0. | In |
min_split_improvementdouble | Minimum relative improvement in squared error reduction for a split to happen. | In |
random_split_pointsboolean | Whether to use random split points for histograms (to pick the best split from). | In |
balance_classesboolean | Balance training data class counts via over/under-sampling (for imbalanced data). | In/Out |
class_sampling_factorsfloat[] | Desired over/under-sampling ratios per class (in lexicographic order). If not specified, sampling factors will be automatically computed to obtain class balance during training. Requires balance_classes. | In/Out |
max_after_balance_sizefloat | Maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes. | In/Out |
max_confusion_matrix_sizeint | Maximum size (# classes) for confusion matrices to be printed in the Logs | In/Out |
max_hit_ratio_kint | Max. number (top K) of predictions to use for hit ratio computation (for multi-class only, 0 to disable) | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
DRFV3
parametersDRFParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
DStackTraceV3
nodestring | Node name | Out |
timelong | Unix epoch time | Out |
thread_tracesstring[] | One trace per thread | Out |
DataInfoFrameV3
frameKey | input frame | In |
interactionsstring[] | interactions | In |
use_allboolean | use all factor levels | In |
standardizeboolean | standardize | In |
interactions_onlyboolean | interactions only returned | In |
resultKey | output frame | Out |
DeepLearningModelOutputV3
weightsKey[] | Frame keys for weight matrices | In |
biasesKey[] | Frame keys for bias vectors | In |
normmuldouble[] | Normalization/Standardization multipliers for numeric predictors | Out |
normsubdouble[] | Normalization/Standardization offsets for numeric predictors | Out |
normrespmuldouble[] | Normalization/Standardization multipliers for numeric response | Out |
normrespsubdouble[] | Normalization/Standardization offsets for numeric response | Out |
catoffsetsint[] | Categorical offsets for one-hot encoding | Out |
variable_importancesTwoDimTable | Variable Importances | Out |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
DeepLearningModelV3
model_idKey | Model key | In/Out |
parametersDeepLearningParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputDeepLearningModelOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
DeepLearningParametersV3
distributionenum | Distribution function | In |
tweedie_powerdouble | Tweedie Power | In |
quantile_alphadouble | Desired quantile for quantile regression (from 0.0 to 1.0) | In |
balance_classesboolean | Balance training data class counts via over/under-sampling (for imbalanced data). | In/Out |
class_sampling_factorsfloat[] | Desired over/under-sampling ratios per class (in lexicographic order). If not specified, sampling factors will be automatically computed to obtain class balance during training. Requires balance_classes. | In/Out |
max_after_balance_sizefloat | Maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes. | In/Out |
max_confusion_matrix_sizeint | Maximum size (# classes) for confusion matrices to be printed in the Logs | In/Out |
max_hit_ratio_kint | Max. number (top K) of predictions to use for hit ratio computation (for multi-class only, 0 to disable) | In/Out |
overwrite_with_best_modelboolean | If enabled, override the final model with the best model found during training | In/Out |
autoencoderboolean | Auto-Encoder | In/Out |
use_all_factor_levelsboolean | Use all factor levels of categorical variables. Otherwise, the first factor level is omitted (without loss of accuracy). Useful for variable importances and auto-enabled for autoencoder. | In/Out |
standardizeboolean | If enabled, automatically standardize the data. If disabled, the user must provide properly scaled input data. | In/Out |
activationenum | Activation function | In/Out |
hiddenint[] | Hidden layer sizes (e.g. 100,100). | In/Out |
epochsdouble | How many times the dataset should be iterated (streamed), can be fractional | In/Out |
train_samples_per_iterationlong | Number of training samples (globally) per MapReduce iteration. Special values are 0: one epoch, -1: all available data (e.g., replicated training data), -2: automatic | In/Out |
target_ratio_comm_to_compdouble | Target ratio of communication overhead to computation. Only for multi-node operation and train_samples_per_iteration=-2 (auto-tuning) | In/Out |
seedlong | Seed for random numbers (affects sampling) - Note: only reproducible when running single threaded | In/Out |
adaptive_rateboolean | Adaptive learning rate | In/Out |
rhodouble | Adaptive learning rate time decay factor (similarity to prior updates) | In/Out |
epsilondouble | Adaptive learning rate smoothing factor (to avoid divisions by zero and allow progress) | In/Out |
ratedouble | Learning rate (higher => less stable, lower => slower convergence) | In/Out |
rate_annealingdouble | Learning rate annealing: rate / (1 + rate_annealing * samples) | In/Out |
rate_decaydouble | Learning rate decay factor between layers (N-th layer: rate*alpha^(N-1)) | In/Out |
momentum_startdouble | Initial momentum at the beginning of training (try 0.5) | In/Out |
momentum_rampdouble | Number of training samples for which momentum increases | In/Out |
momentum_stabledouble | Final momentum after the ramp is over (try 0.99) | In/Out |
nesterov_accelerated_gradientboolean | Use Nesterov accelerated gradient (recommended) | In/Out |
input_dropout_ratiodouble | Input layer dropout ratio (can improve generalization, try 0.1 or 0.2) | In/Out |
hidden_dropout_ratiosdouble[] | Hidden layer dropout ratios (can improve generalization), specify one value per hidden layer, defaults to 0.5 | In/Out |
l1double | L1 regularization (can add stability and improve generalization, causes many weights to become 0) | In/Out |
l2double | L2 regularization (can add stability and improve generalization, causes many weights to be small | In/Out |
max_w2float | Constraint for squared sum of incoming weights per unit (e.g. for Rectifier) | In/Out |
initial_weight_distributionenum | Initial Weight Distribution | In/Out |
initial_weight_scaledouble | Uniform: -value…value, Normal: stddev) | In/Out |
initial_weightsKey[] | A list of H2OFrame ids to initialize the weight matrices of this model with. | In/Out |
initial_biasesKey[] | A list of H2OFrame ids to initialize the bias vectors of this model with. | In/Out |
lossenum | Loss function | In/Out |
score_intervaldouble | Shortest time interval (in secs) between model scoring | In/Out |
score_training_sampleslong | Number of training set samples for scoring (0 for all) | In/Out |
score_validation_sampleslong | Number of validation set samples for scoring (0 for all) | In/Out |
score_duty_cycledouble | Maximum duty cycle fraction for scoring (lower: more training, higher: more scoring). | In/Out |
classification_stopdouble | Stopping criterion for classification error fraction on training data (-1 to disable) | In/Out |
regression_stopdouble | Stopping criterion for regression error (MSE) on training data (-1 to disable) | In/Out |
quiet_modeboolean | Enable quiet mode for less output to standard output | In/Out |
score_validation_samplingenum | Method used to sample validation dataset for scoring | In/Out |
diagnosticsboolean | Enable diagnostics for hidden layers | In/Out |
variable_importancesboolean | Compute variable importances for input features (Gedeon method) - can be slow for large networks | In/Out |
fast_modeboolean | Enable fast mode (minor approximation in back-propagation) | In/Out |
force_load_balanceboolean | Force extra load balancing to increase training speed for small datasets (to keep all cores busy) | In/Out |
replicate_training_databoolean | Replicate the entire training dataset onto every node for faster training on small datasets | In/Out |
single_node_modeboolean | Run on a single node for fine-tuning of model parameters | In/Out |
shuffle_training_databoolean | Enable shuffling of training data (recommended if training data is replicated and train_samples_per_iteration is close to #nodes x #rows, of if using balance_classes) | In/Out |
missing_values_handlingenum | Handling of missing values. Either Skip or MeanImputation. | In/Out |
sparseboolean | Sparse data handling (more efficient for data with lots of 0 values). | In/Out |
col_majorboolean | Use a column major weight matrix for input layer. Can speed up forward propagation, but might slow down backpropagation (Deprecated). | In/Out |
average_activationdouble | Average activation for sparse auto-encoder (Experimental) | In/Out |
sparsity_betadouble | Sparsity regularization (Experimental) | In/Out |
max_categorical_featuresint | Max. number of categorical features, enforced via hashing (Experimental) | In/Out |
reproducibleboolean | Force reproducibility on small data (will be slow - only uses 1 thread) | In/Out |
export_weights_and_biasesboolean | Whether to export Neural Network weights and biases to H2O Frames | In/Out |
mini_batch_sizeint | Mini-batch size (smaller leads to better fit, larger can speed up and generalize better) | In/Out |
elastic_averagingboolean | Elastic averaging between compute nodes can improve distributed model convergence (Experimental) | In/Out |
elastic_averaging_moving_ratedouble | Elastic averaging moving rate (only if elastic averaging is enabled). | In/Out |
elastic_averaging_regularizationdouble | Elastic averaging regularization strength (only if elastic averaging is enabled). | In/Out |
pretrained_autoencoderKey | Pretrained autoencoder model to initialize this model with. | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
DeepLearningV3
parametersDeepLearningParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
DownloadDataV3
frame_idKey | Frame to download | In |
hex_stringboolean | Emit double values in a machine readable lossless format with Double.toHexString(). | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
csvstring | CSV Stream | Out |
filenamestring | Suggested Filename | Out |
EventV3
datestring | Time when the event was recorded. Format is hh:mm:ss:ms | In |
nanoslong | Time in nanos | In |
typeenum | type of recorded event | In |
ExampleModelOutputV3
iterationsint | Iterations executed | In |
maxsdouble[] | (No description available) | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
ExampleModelV3
model_idKey | Model key | In/Out |
parametersExampleParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputExampleOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
ExampleParametersV3
max_iterationsint | Maximum training iterations. | In |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
ExampleV3
parametersExampleParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
FieldMetadataBase
schema_namestring | Schema name for this field, if it is_schema, or the name of the enum, if it’s an enum. | In |
namestring | Field name in the Schema | Out |
typestring | Type for this field | Out |
is_schemaboolean | Type for this field is itself a Schema. | Out |
valuePolymorphic | Value for this field | Out |
helpstring | A short help description to appear alongside the field in a UI | Out |
labelstring | The label that should be displayed for the field if the name is insufficient | Out |
requiredboolean | Is this field required, or is the default value generally sufficient? | Out |
levelenum | How important is this field? The web UI uses the level to do a slow reveal of the parameters | Out |
directionenum | Is this field an input, output or inout? | Out |
is_inheritedboolean | Is the field inherited from the parent schema? | Out |
is_gridableboolean | Is the field gridable (i.e., it can be used in grid call) | Out |
valuesstring[] | For enum-type fields the allowed values are specified using the values annotation; this is used in UIs to tell the user the allowed values, and for validation | Out |
jsonboolean | Should this field be rendered in the JSON representation? | Out |
is_member_of_framesstring[] | For Vec-type fields this is the set of other Vec-type fields which must contain mutually exclusive values; for example, for a SupervisedModel the response_column must be mutually exclusive with the weights_column | Out |
is_mutually_exclusive_withstring[] | For Vec-type fields this is the set of Frame-type fields which must contain the named column; for example, for a SupervisedModel the response_column must be in both the training_frame and (if it’s set) the validation_frame | Out |
FieldMetadataV3
schema_namestring | Schema name for this field, if it is_schema, or the name of the enum, if it’s an enum. | In |
namestring | Field name in the Schema | Out |
typestring | Type for this field | Out |
is_schemaboolean | Type for this field is itself a Schema. | Out |
valuePolymorphic | Value for this field | Out |
helpstring | A short help description to appear alongside the field in a UI | Out |
labelstring | The label that should be displayed for the field if the name is insufficient | Out |
requiredboolean | Is this field required, or is the default value generally sufficient? | Out |
levelenum | How important is this field? The web UI uses the level to do a slow reveal of the parameters | Out |
directionenum | Is this field an input, output or inout? | Out |
is_inheritedboolean | Is the field inherited from the parent schema? | Out |
is_gridableboolean | Is the field gridable (i.e., it can be used in grid call) | Out |
valuesstring[] | For enum-type fields the allowed values are specified using the values annotation; this is used in UIs to tell the user the allowed values, and for validation | Out |
jsonboolean | Should this field be rendered in the JSON representation? | Out |
is_member_of_framesstring[] | For Vec-type fields this is the set of other Vec-type fields which must contain mutually exclusive values; for example, for a SupervisedModel the response_column must be mutually exclusive with the weights_column | Out |
is_mutually_exclusive_withstring[] | For Vec-type fields this is the set of Frame-type fields which must contain the named column; for example, for a SupervisedModel the response_column must be in both the training_frame and (if it’s set) the validation_frame | Out |
FindV3
keyFrame | Frame to search | In |
columnstring | Column, or null for all | In |
rowlong | Starting row for search | In |
matchstring | Value to search for; leave blank for a search for missing values | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
prevlong | previous row with matching value, or -1 | Out |
nextlong | next row with matching value, or -1 | Out |
FrameBase
frame_idKey | Frame ID | In/Out |
byte_sizelong | Total data size in bytes | Out |
is_textboolean | Is this Frame raw unparsed data? | Out |
FrameKeyV3
namestring | Name (string representation) for this Key. | In/Out |
typestring | Name (string representation) for the type of Keyed this Key points to. | In/Out |
URLstring | URL for the resource that this Key points to, if one exists. | In/Out |
FrameSynopsisV3
frame_idKey | Frame ID | In/Out |
rowslong | Number of rows in the Frame | Out |
columnslong | Number of columns in the Frame | Out |
byte_sizelong | Total data size in bytes | Out |
is_textboolean | Is this Frame raw unparsed data? | Out |
FrameV3
row_offsetlong | Row offset to display | In |
row_countint | Number of rows to display | In/Out |
column_offsetint | Column offset to return | In/Out |
column_countint | Number of columns to return | In/Out |
total_column_countint | Total number of columns in the Frame | In/Out |
frame_idKey | Frame ID | In/Out |
checksumlong | checksum | Out |
rowslong | Number of rows in the Frame | Out |
num_columnslong | Number of columns in the Frame | Out |
default_percentilesdouble[] | Default percentiles, from 0 to 1 | Out |
columnsVec[] | Columns in the Frame | Out |
compatible_modelsstring[] | Compatible models, if requested | Out |
chunk_summaryTwoDimTable | Chunk summary | Out |
distribution_summaryTwoDimTable | Distribution summary | Out |
byte_sizelong | Total data size in bytes | Out |
is_textboolean | Is this Frame raw unparsed data? | Out |
FramesBase
frame_idKey | Name of Frame of interest | In |
columnstring | Name of column of interest | In |
find_compatible_modelsboolean | Find and return compatible models? | In |
pathstring | File output path | In |
forceboolean | Overwrite existing file | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
row_offsetlong | Row offset to return | In/Out |
row_countint | Number of rows to return | In/Out |
column_offsetint | Column offset to return | In/Out |
column_countint | Number of columns to return | In/Out |
jobJob | Job for export file | Out |
framesIced[] | Frames | Out |
compatible_modelsModel[] | Compatible models | Out |
domainstring[][] | Domains | Out |
FramesV3
frame_idKey | Name of Frame of interest | In |
columnstring | Name of column of interest | In |
find_compatible_modelsboolean | Find and return compatible models? | In |
pathstring | File output path | In |
forceboolean | Overwrite existing file | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
row_offsetlong | Row offset to return | In/Out |
row_countint | Number of rows to return | In/Out |
column_offsetint | Column offset to return | In/Out |
column_countint | Number of columns to return | In/Out |
jobJob | Job for export file | Out |
framesIced[] | Frames | Out |
compatible_modelsModel[] | Compatible models | Out |
domainstring[][] | Domains | Out |
GBMModelOutputV3
variable_importancesTwoDimTable | Variable Importances | Out |
init_fdouble | The Intercept term, the initial model function value to which trees make adjustments | Out |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
GBMModelV3
model_idKey | Model key | In/Out |
parametersGBMParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputGBMOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
GBMParametersV3
learn_ratedouble | Learning rate (from 0.0 to 1.0) | In |
learn_rate_annealingdouble | Scale down the learning rate by this factor after each tree | In |
distributionenum | Distribution function | In |
quantile_alphadouble | Desired quantile for quantile regression (from 0.0 to 1.0) | In |
tweedie_powerdouble | Tweedie Power (between 1 and 2) | In |
col_sample_ratedouble | Column sample rate (from 0.0 to 1.0) | In |
max_abs_leafnode_preddouble | Maximum absolute value of a leaf node prediction | In |
ntreesint | Number of trees. | In |
max_depthint | Maximum tree depth. | In |
min_rowsdouble | Fewest allowed (weighted) observations in a leaf (in R called ‘nodesize’). | In |
nbinsint | For numerical columns (real/int), build a histogram of (at least) this many bins, then split at the best point | In |
nbins_top_levelint | For numerical columns (real/int), build a histogram of (at most) this many bins at the root level, then decrease by factor of two per level | In |
nbins_catsint | For categorical columns (factors), build a histogram of this many bins, then split at the best point. Higher values can lead to more overfitting. | In |
r2_stoppingdouble | Stop making trees when the R^2 metric equals or exceeds this | In |
seedlong | Seed for pseudo random number generator (if applicable) | In |
build_tree_one_nodeboolean | Run on one node only; no network overhead but fewer cpus used. Suitable for small datasets. | In |
sample_ratedouble | Row sample rate per tree (from 0.0 to 1.0) | In |
sample_rate_per_classdouble[] | Row sample rate per tree per class (from 0.0 to 1.0) | In |
col_sample_rate_per_treedouble | Column sample rate per tree (from 0.0 to 1.0) | In |
col_sample_rate_change_per_leveldouble | Relative change of the column sampling rate for every level (from 0.0 to 2.0) | In |
score_tree_intervalint | Score the model after every so many trees. Disabled if set to 0. | In |
min_split_improvementdouble | Minimum relative improvement in squared error reduction for a split to happen. | In |
random_split_pointsboolean | Whether to use random split points for histograms (to pick the best split from). | In |
balance_classesboolean | Balance training data class counts via over/under-sampling (for imbalanced data). | In/Out |
class_sampling_factorsfloat[] | Desired over/under-sampling ratios per class (in lexicographic order). If not specified, sampling factors will be automatically computed to obtain class balance during training. Requires balance_classes. | In/Out |
max_after_balance_sizefloat | Maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes. | In/Out |
max_confusion_matrix_sizeint | Maximum size (# classes) for confusion matrices to be printed in the Logs | In/Out |
max_hit_ratio_kint | Max. number (top K) of predictions to use for hit ratio computation (for multi-class only, 0 to disable) | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
GBMV3
parametersGBMParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
GLMModelOutputV3
coefficients_tableTwoDimTable | Table of Coefficients | In |
standardized_coefficient_magnitudesTwoDimTable | Standardized Coefficient Magnitudes | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
GLMModelV3
model_idKey | Model key | In/Out |
parametersGLMParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputGLMOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
GLMParametersV3
familyenum | Family. Use binomial for classification with logistic regression, others are for regression problems. | In |
tweedie_variance_powerdouble | Tweedie variance power | In |
tweedie_link_powerdouble | Tweedie link power | In |
solverenum | AUTO will set the solver based on given data and the other parameters. IRLSM is fast on on problems with small number of predictors and for lambda-search with L1 penalty, L_BFGS scales better for datasets with many columns. Coordinate descent is experimental (beta). | In |
alphadouble[] | distribution of regularization between L1 and L2. | In |
lambdadouble[] | regularization strength | In |
lambda_searchboolean | use lambda search starting at lambda max, given lambda is then interpreted as lambda min | In |
nlambdasint | number of lambdas to be used in a search | In |
standardizeboolean | Standardize numeric columns to have zero mean and unit variance | In |
non_negativeboolean | Restrict coefficients (not intercept) to be non-negative | In |
max_iterationsint | Maximum number of iterations | In |
beta_epsilondouble | converge if beta changes less (using L-infinity norm) than beta esilon, ONLY applies to IRLSM solver | In |
objective_epsilondouble | converge if objective value changes less than this | In |
gradient_epsilondouble | converge if objective changes less (using L-infinity norm) than this, ONLY applies to L-BFGS solver | In |
obj_regdouble | likelihood divider in objective value computation, default is 1/nobs | In |
linkenum | (No description available) | In |
interceptboolean | include constant term in the model | In |
priordouble | prior probability for y==1. To be used only for logistic regression iff the data has been sampled and the mean of response does not reflect reality. | In |
lambda_min_ratiodouble | min lambda used in lambda search, specified as a ratio of lambda_max | In |
beta_constraintsKey | beta constraints | In |
max_active_predictorsint | Maximum number of active predictors during computation. Use as a stopping criterium to prevent expensive model building with many predictors. | In |
interactionsstring[] | A list of predictor column indices to interact. All pairwise combinations will be computed for the list. | In |
compute_p_valuesboolean | request p-values computation, p-values work only with IRLSM solver and no regularization | In |
remove_collinear_columnsboolean | in case of linearly dependent columns remove some of the dependent columns | In |
missing_values_handlingenum | Handling of missing values. Either Skip or MeanImputation. | In/Out |
balance_classesboolean | Balance training data class counts via over/under-sampling (for imbalanced data). | In/Out |
class_sampling_factorsfloat[] | Desired over/under-sampling ratios per class (in lexicographic order). If not specified, sampling factors will be automatically computed to obtain class balance during training. Requires balance_classes. | In/Out |
max_after_balance_sizefloat | Maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes. | In/Out |
max_confusion_matrix_sizeint | Maximum size (# classes) for confusion matrices to be printed in the Logs | In/Out |
max_hit_ratio_kint | Max. number (top K) of predictions to use for hit ratio computation (for multi-class only, 0 to disable) | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
GLMV3
parametersGLMParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
GLRMModelOutputV3
iterationsint | Iterations executed | In |
updatesint | Updates executed | In |
objectivedouble | Objective value | In |
avg_change_objdouble | Average change in objective value on final iteration | In |
step_sizedouble | Final step size | In |
archetypesTwoDimTable | Mapping from lower dimensional k-space to training features | In |
singular_valsdouble[] | Singular values of XY matrix | In |
eigenvectorsTwoDimTable | Eigenvectors of XY matrix | In |
representation_namestring | Frame key name for X matrix | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
GLRMModelV3
model_idKey | Model key | In/Out |
parametersGLRMParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputGLRMOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
GLRMParametersV3
transformenum | Transformation of training data | In |
kint | Rank of matrix approximation | In |
lossenum | Numeric loss function | In |
multi_lossenum | Categorical loss function | In |
loss_by_colenum[] | Loss function by column (override) | In |
loss_by_col_idxint[] | Loss function by column index (override) | In |
periodint | Length of period (only used with periodic loss function) | In |
regularization_xenum | Regularization function for X matrix | In |
regularization_yenum | Regularization function for Y matrix | In |
gamma_xdouble | Regularization weight on X matrix | In |
gamma_ydouble | Regularization weight on Y matrix | In |
max_iterationsint | Maximum number of iterations | In |
max_updatesint | Maximum number of updates | In |
init_step_sizedouble | Initial step size | In |
min_step_sizedouble | Minimum step size | In |
seedlong | RNG seed for initialization | In |
initenum | Initialization mode | In |
svd_methodenum | Method for computing SVD during initialization (Caution: Power and Randomized are currently experimental and unstable) | In |
user_yKey | User-specified initial Y | In |
user_xKey | User-specified initial X | In |
loading_namestring | Frame key to save resulting X | In |
expand_user_yboolean | Expand categorical columns in user-specified initial Y | In |
impute_originalboolean | Reconstruct original training data by reversing transform | In |
recover_svdboolean | Recover singular values and eigenvectors of XY | In |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
GLRMV3
parametersGLRMParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
GarbageCollectV3
(No fields)
GrepModelOutputV3
matchesstring[] | Matching strings | In |
offsetslong[] | Byte offsets of matches | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
GrepModelV3
model_idKey | Model key | In/Out |
parametersGrepParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputGrepOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
GrepParametersV3
regexstring | regex | In |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
GrepV3
parametersGrepParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
GridKeyV3
namestring | Name (string representation) for this Key. | In/Out |
typestring | Name (string representation) for the type of Keyed this Key points to. | In/Out |
URLstring | URL for the resource that this Key points to, if one exists. | In/Out |
GridSchemaV99
grid_idKey | Grid id | In |
model_idsKey | Model IDs built by a grid search | In |
sort_bystring | Model performance metric to sort by. Examples: logloss, residual_deviance, mse, auc, r2, f1, recall, precision, accuracy, mcc, err, err_count, lift_top_group, max_per_class_error | In/Out |
decreasingboolean | Specify whether sort order should be decreasing. | In/Out |
hyper_namesstring[] | Used hyper parameters. | Out |
failed_paramsParameters[] | List of failed parameters | Out |
failure_detailsstring[] | List of detailed failure messages | Out |
failure_stack_tracesstring[] | List of detailed failure stack traces | Out |
failed_raw_paramsstring[][] | List of raw parameters causing model building failure | Out |
training_metricsModelMetrics[] | Training model metrics for the returned models; only returned if sort_by is set | Out |
validation_metricsModelMetrics[] | Validation model metrics for the returned models; only returned if sort_by is set | Out |
cross_validation_metricsModelMetrics[] | Cross validation model metrics for the returned models; only returned if sort_by is set | Out |
cross_validation_metrics_summaryTwoDimTable[] | Cross validation model metrics summary for the returned models; only returned if sort_by is set | Out |
summary_tableTwoDimTable | Summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
GridSearchSchema
parametersParameters | Basic model builder parameters. | In |
hyper_parametersMap | Grid search parameters. | In/Out |
grid_idKey | Destination id for this grid; auto-generated if not specified. | In/Out |
search_criteriaHyperSpaceSearchCriteria | Hyperparameter search criteria, including strategy and early stopping directives. If it is not given, exhaustive Cartesian is used. | In/Out |
total_modelsint | Number of all models generated by grid search. | Out |
jobJob | Job Key. | Out |
GridsV99
gridsGrid[] | Grids | Out |
H2OErrorV3
timestamplong | Milliseconds since the epoch for the time that this H2OError instance was created. Generally this is a short time since the underlying error ocurred. | Out |
error_urlstring | Error url | Out |
msgstring | Message intended for the end user (a data scientist). | Out |
dev_msgstring | Potentially more detailed message intended for a developer (e.g. a front end engineer or someone designing a language binding). | Out |
http_statusint | HTTP status code for this error. | Out |
valuesMap | Any values that are relevant to reporting or handling this error. Examples are a key name if the error is on a key, or a field name and object name if it’s on a specific field. | Out |
exception_typestring | Exception type, if any. | Out |
exception_msgstring | Raw exception message, if any. | Out |
stacktracestring[] | Stacktrace, if any. | Out |
H2OModelBuilderErrorV3
parametersParameters | Model builder parameters. | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
timestamplong | Milliseconds since the epoch for the time that this H2OError instance was created. Generally this is a short time since the underlying error ocurred. | Out |
error_urlstring | Error url | Out |
msgstring | Message intended for the end user (a data scientist). | Out |
dev_msgstring | Potentially more detailed message intended for a developer (e.g. a front end engineer or someone designing a language binding). | Out |
http_statusint | HTTP status code for this error. | Out |
valuesMap | Any values that are relevant to reporting or handling this error. Examples are a key name if the error is on a key, or a field name and object name if it’s on a specific field. | Out |
exception_typestring | Exception type, if any. | Out |
exception_msgstring | Raw exception message, if any. | Out |
stacktracestring[] | Stacktrace, if any. | Out |
HeartBeatEvent
sendsint | number of sent heartbeats | In |
recvsint | number of received heartbeats | In |
datestring | Time when the event was recorded. Format is hh:mm:ss:ms | In |
nanoslong | Time in nanos | In |
typeenum | type of recorded event | In |
HyperSpaceSearchCriteriaV99
strategyenum | Hyperparameter space search strategy. | In/Out |
IOEvent
io_flavorstring | flavor of the recorded io (ice/hdfs/…) | In |
nodestring | node where this io event happened | In |
datastring | data info | In |
datestring | Time when the event was recorded. Format is hh:mm:ss:ms | In |
nanoslong | Time in nanos | In |
typeenum | type of recorded event | In |
ImportFilesV3
pathstring | path | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
filesstring[] | files | Out |
destination_framesstring[] | names | Out |
failsstring[] | fails | Out |
delsstring[] | dels | Out |
ImportSQLTableV99
connection_urlstring | connection_url | In |
tablestring | table | In |
select_querystring | select_query | In |
usernamestring | username | In |
passwordstring | password | In |
columnsstring | columns | In |
optimizeboolean | optimize | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
InitIDV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
session_keystring | Session ID | Out |
InteractionV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
destKey | destination key | In/Out |
source_frameKey | Input data frame | In/Out |
factor_columnsstring[] | Factor columns | In/Out |
pairwiseboolean | Whether to create pairwise quadratic interactions between factors (otherwise create one higher-order interaction). Only applicable if there are 3 or more factors. | In/Out |
max_factorsint | Max. number of factor levels in pair-wise interaction terms (if enforced, one extra catch-all factor will be made) | In/Out |
min_occurrenceint | Min. occurrence threshold for factor levels in pair-wise interaction terms | In/Out |
IoStatsEntry
backendstring | Back end type | Out |
store_countlong | Number of store events | Out |
store_byteslong | Cumulative stored bytes | Out |
delete_countlong | Number of delete events | Out |
load_countlong | Number of load events | Out |
load_byteslong | Cumulative loaded bytes | Out |
JStackV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
tracesDStackTrace[] | Stacktraces | Out |
JobKeyV3
namestring | Name (string representation) for this Key. | In/Out |
typestring | Name (string representation) for the type of Keyed this Key points to. | In/Out |
URLstring | URL for the resource that this Key points to, if one exists. | In/Out |
JobV3
keyKey | Job Key | In |
descriptionstring | Job description | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
destKey | destination key | In/Out |
statusstring | job status | Out |
progressfloat | progress, from 0 to 1 | Out |
progress_msgstring | current progress status description | Out |
start_timelong | Start time | Out |
mseclong | Runtime in milliseconds | Out |
warningsstring[] | exception | Out |
exceptionstring | exception | Out |
stacktracestring | stacktrace | Out |
ready_for_viewboolean | ready for view | Out |
JobsV3
job_idKey | Optional Job identifier | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
jobsJob[] | jobs | Out |
KMeansModelOutputV3
centersTwoDimTable | Cluster Centers[k][features] | In |
centers_stdTwoDimTable | Cluster Centers[k][features] on Standardized Data | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
KMeansModelV3
model_idKey | Model key | In/Out |
parametersKMeansParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputKMeansOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
KMeansParametersV3
user_pointsKey | User-specified points | In |
max_iterationsint | Maximum training iterations | In |
standardizeboolean | Standardize columns | In |
seedlong | RNG Seed | In |
initenum | Initialization mode | In |
kint | Number of clusters | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
KMeansV3
parametersKMeansParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
KeyV3
namestring | Name (string representation) for this Key. | In/Out |
typestring | Name (string representation) for the type of Keyed this Key points to. | In/Out |
URLstring | URL for the resource that this Key points to, if one exists. | In/Out |
KillMinus3V3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
LogAndEchoV3
messagestring | Message to be Logged and Echoed | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
LogsV3
nodeidxint | Index of node to query ticks for (0-based). -1 means current node. | In |
namestring | Which specific log file to read from the log file directory. If left unspecified, the system chooses a default for you. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
logstring | Content of log file | Out |
MakeGLMModelV3
modelKey | source model | In |
destKey | destination key | In |
namesstring[] | coefficient names | In |
betadouble[] | new glm coefficients | In |
thresholdfloat | decision threshold for label-generation | In |
MetadataBase
numint | Number for specifying an endpoint | In |
http_methodstring | HTTP method (GET, POST, DELETE) if fetching by path | In |
pathstring | Path for specifying an endpoint | In |
classnamestring | Class name, for fetching docs for a schema (DEPRECATED) | In |
schemanamestring | Schema name (e.g., DocsV1), for fetching docs for a schema | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
routesRoute[] | List of endpoint routes | Out |
schemasSchemaMetadata[] | List of schemas | Out |
markdownstring | Table of Contents Markdown | Out |
MetadataV3
numint | Number for specifying an endpoint | In |
http_methodstring | HTTP method (GET, POST, DELETE) if fetching by path | In |
pathstring | Path for specifying an endpoint | In |
classnamestring | Class name, for fetching docs for a schema (DEPRECATED) | In |
schemanamestring | Schema name (e.g., DocsV1), for fetching docs for a schema | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
routesRoute[] | List of endpoint routes | Out |
schemasSchemaMetadata[] | List of schemas | Out |
markdownstring | Table of Contents Markdown | Out |
MissingInserterV3
datasetKey | dataset | In |
fractiondouble | Fraction of data to replace with a missing value | In |
seedlong | Seed | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
ModelBuilderSchema
parametersParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
ModelBuilderV3
parametersParameters | Model builder parameters. | Out |
messagesValidationMessage[] | Info, warning and error messages; NOTE: can be appended to while the Job is running | Out |
error_countint | Count of error messages | Out |
ModelBuildersBase
algostring | Algo of ModelBuilder of interest | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
model_buildersMap | ModelBuilders | Out |
ModelBuildersV3
algostring | Algo of ModelBuilder of interest | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
model_buildersMap | ModelBuilders | Out |
ModelExportV3
model_idKey | Name of Model of interest | In |
dirstring | Destination file (hdfs, s3, local) | In |
forceboolean | Overwrite destination file in case it exists or throw exception if set to false. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
ModelIdV3
model_idstring | Model ID | Out |
ModelImportV3
model_idKey | Save imported model under given key into DKV. | In |
dirstring | Source directory (hdfs, s3, local) containing serialized model | In |
forceboolean | Override existing model in case it exists or throw exception if set to false | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
ModelKeyV3
namestring | Name (string representation) for this Key. | In/Out |
typestring | Name (string representation) for the type of Keyed this Key points to. | In/Out |
URLstring | URL for the resource that this Key points to, if one exists. | In/Out |
ModelMetricsAutoEncoderV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsBase
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsBinomialGLMV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
residual_deviancedouble | residual deviance | Out |
null_deviancedouble | null deviance | Out |
AICdouble | AIC | Out |
null_degrees_of_freedomlong | null DOF | Out |
residual_degrees_of_freedomlong | residual DOF | Out |
r2double | The R^2 for this scoring run. | Out |
loglossdouble | The logarithmic loss for this scoring run. | Out |
AUCdouble | The AUC for this scoring run. | Out |
Ginidouble | The Gini score for this scoring run. | Out |
domainstring[] | The class labels of the response. | Out |
thresholds_and_metric_scoresTwoDimTable | The Metrics for various thresholds. | Out |
max_criteria_and_metric_scoresTwoDimTable | The Metrics for various criteria. | Out |
gains_lift_tableTwoDimTable | Gains and Lift table. | Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsBinomialV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
r2double | The R^2 for this scoring run. | Out |
loglossdouble | The logarithmic loss for this scoring run. | Out |
AUCdouble | The AUC for this scoring run. | Out |
Ginidouble | The Gini score for this scoring run. | Out |
domainstring[] | The class labels of the response. | Out |
thresholds_and_metric_scoresTwoDimTable | The Metrics for various thresholds. | Out |
max_criteria_and_metric_scoresTwoDimTable | The Metrics for various criteria. | Out |
gains_lift_tableTwoDimTable | Gains and Lift table. | Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsClusteringV3
tot_withinssdouble | Within Cluster Sum of Square Error | In |
totssdouble | Total Sum of Square Error to Grand Mean | In |
betweenssdouble | Between Cluster Sum of Square Error | In |
centroid_statsTwoDimTable | Centroid Statistics | In |
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsGLRMV99
numerrdouble | Sum of Squared Error (Numeric Cols) | In |
caterrdouble | Misclassification Error (Categorical Cols) | In |
numcntlong | Number of Non-Missing Numeric Values | In |
catcntlong | Number of Non-Missing Categorical Values | In |
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsListSchemaV3
modelKey | Key of Model of interest (optional) | In |
frameKey | Key of Frame of interest (optional) | In |
reconstruction_errorboolean | Compute reconstruction error (optional, only for Deep Learning AutoEncoder models) | In |
reconstruction_error_per_featureboolean | Compute reconstruction error per feature (optional, only for Deep Learning AutoEncoder models) | In |
deep_features_hidden_layerint | Extract Deep Features for given hidden layer (optional, only for Deep Learning models) | In |
reconstruct_trainboolean | Reconstruct original training frame (optional, only for GLRM models) | In |
project_archetypesboolean | Project GLRM archetypes back into original feature space (optional, only for GLRM models) | In |
reverse_transformboolean | Reverse transformation applied during training to model output (optional, only for GLRM models) | In |
leaf_node_assignmentboolean | Return the leaf node assignment (optional, only for DRF/GBM models) | In |
exemplar_indexint | Retrieve all members for a given exemplar (optional, only for Aggregator models) | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
predictions_frameKey | Key of predictions frame, if predictions are requested (optional) | In/Out |
model_metricsModelMetrics[] | ModelMetrics | Out |
ModelMetricsMultinomialGLMV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
residual_deviancedouble | residual deviance | Out |
null_deviancedouble | null deviance | Out |
AICdouble | AIC | Out |
null_degrees_of_freedomlong | null DOF | Out |
residual_degrees_of_freedomlong | residual DOF | Out |
r2double | The R^2 for this scoring run. | Out |
hit_ratio_tableTwoDimTable | The hit ratio table for this scoring run. | Out |
cmConfusionMatrix | The ConfusionMatrix object for this scoring run. | Out |
loglossdouble | The logarithmic loss for this scoring run. | Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsMultinomialV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
r2double | The R^2 for this scoring run. | Out |
hit_ratio_tableTwoDimTable | The hit ratio table for this scoring run. | Out |
cmConfusionMatrix | The ConfusionMatrix object for this scoring run. | Out |
loglossdouble | The logarithmic loss for this scoring run. | Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsPCAV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsRegressionGLMV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
residual_deviancedouble | residual deviance | Out |
null_deviancedouble | null deviance | Out |
AICdouble | AIC | Out |
null_degrees_of_freedomlong | null DOF | Out |
residual_degrees_of_freedomlong | residual DOF | Out |
r2double | The R^2 for this scoring run. | Out |
mean_residual_deviancedouble | The mean residual deviance for this scoring run. | Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsRegressionV3
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
r2double | The R^2 for this scoring run. | Out |
mean_residual_deviancedouble | The mean residual deviance for this scoring run. | Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelMetricsSVDV99
modelKey | The model used for this scoring run. | In/Out |
model_checksumlong | The checksum for the model used for this scoring run. | In/Out |
frameKey | The frame used for this scoring run. | In/Out |
frame_checksumlong | The checksum for the frame used for this scoring run. | In/Out |
descriptionstring | Optional description for this scoring run (to note out-of-bag, sampled data, etc.) | Out |
model_categoryenum | The category (e.g., Clustering) for the model used for this scoring run. | Out |
scoring_timelong | The time in mS since the epoch for the start of this scoring run. | Out |
predictionsFrame | Predictions Frame. | Out |
MSEdouble | The Mean Squared Error of the prediction for this scoring run. | Out |
ModelOutputSchema
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
ModelParameterSchemaV3
is_member_of_framesstring[] | For Vec-type fields this is the set of other Vec-type fields which must contain mutually exclusive values; for example, for a SupervisedModel the response_column must be mutually exclusive with the weights_column | In |
is_mutually_exclusive_withstring[] | For Vec-type fields this is the set of Frame-type fields which must contain the named column; for example, for a SupervisedModel the response_column must be in both the training_frame and (if it’s set) the validation_frame | In |
namestring | name in the JSON, e.g. “lambda” | Out |
labelstring | label in the UI, e.g. “lambda” | Out |
helpstring | help for the UI, e.g. “regularization multiplier, typically used for foo bar baz etc.” | Out |
requiredboolean | the field is required | Out |
typestring | Java type, e.g. “double” | Out |
default_valuePolymorphic | default value, e.g. 1 | Out |
actual_valuePolymorphic | actual value as set by the user and / or modified by the ModelBuilder, e.g., 10 | Out |
levelstring | the importance of the parameter, used by the UI, e.g. “critical”, “extended” or “expert” | Out |
valuesstring[] | list of valid values for use by the front-end | Out |
gridableboolean | Parameter can be used in grid call | Out |
ModelParametersSchema
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
ModelSchema
model_idKey | Model key | In/Out |
parametersParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
ModelSchemaBase
model_idKey | Model key | In/Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
ModelSynopsisV3
model_idKey | Model key | In/Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
ModelsBase
model_idKey | Name of Model of interest | In |
previewboolean | Return potentially abridged model suitable for viewing in a browser | In |
find_compatible_framesboolean | Find and return compatible frames? | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
modelsIced[] | Models | Out |
compatible_framesFrame[] | Compatible frames | Out |
ModelsV3
model_idKey | Name of Model of interest | In |
previewboolean | Return potentially abridged model suitable for viewing in a browser | In |
find_compatible_framesboolean | Find and return compatible frames? | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
modelsIced[] | Models | Out |
compatible_framesFrame[] | Compatible frames | Out |
NaiveBayesModelOutputV3
levelsstring[] | Categorical levels of the response | In |
aprioriTwoDimTable | A-priori probabilities of the response | In |
pcondTwoDimTable[] | Conditional probabilities of the predictors | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
NaiveBayesModelV3
model_idKey | Model key | In/Out |
parametersNaiveBayesParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputNaiveBayesOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
NaiveBayesParametersV3
laplacedouble | Laplace smoothing parameter | In |
min_sdevdouble | Min. standard deviation to use for observations with not enough data | In |
eps_sdevdouble | Cutoff below which standard deviation is replaced with min_sdev | In |
min_probdouble | Min. probability to use for observations with not enough data | In |
eps_probdouble | Cutoff below which probability is replaced with min_prob | In |
compute_metricsboolean | Compute metrics on training data | In |
balance_classesboolean | Balance training data class counts via over/under-sampling (for imbalanced data). | In/Out |
class_sampling_factorsfloat[] | Desired over/under-sampling ratios per class (in lexicographic order). If not specified, sampling factors will be automatically computed to obtain class balance during training. Requires balance_classes. | In/Out |
max_after_balance_sizefloat | Maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes. | In/Out |
max_confusion_matrix_sizeint | Maximum size (# classes) for confusion matrices to be printed in the Logs | In/Out |
max_hit_ratio_kint | Max. number (top K) of predictions to use for hit ratio computation (for multi-class only, 0 to disable) | In/Out |
seedlong | Seed for pseudo random number generator (only used for cross-validation and fold_assignment=”Random” or “AUTO”) | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
NaiveBayesV3
parametersNaiveBayesParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
NetworkBenchV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
resultsTwoDimTable[] | NetworkBenchResults | Out |
NetworkEvent
is_sendboolean | Boolean flag distinguishing between sends (true) and receives(false) | In |
protocolstring | network protocol (UDP/TCP) | In |
msg_typestring | UDP type (exec,ack, ackack,… | In |
fromstring | Sending node | In |
tostring | Receiving node | In |
datastring | Pretty print of the first few bytes of the msg payload. Contains class name for tasks. | In |
datestring | Time when the event was recorded. Format is hh:mm:ss:ms | In |
nanoslong | Time in nanos | In |
typeenum | type of recorded event | In |
NetworkTestV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
microseconds_collectivedouble[] | Collective broadcast/reduce times in microseconds (for each message size) | Out |
bandwidths_collectivedouble[] | Collective bandwidths in Bytes/sec (for each message size, for each node) | Out |
microsecondsdouble[][] | Round-trip times in microseconds (for each message size, for each node) | Out |
bandwidthsdouble[][] | Bi-directional bandwidths in Bytes/sec (for each message size, for each node) | Out |
nodesstring[] | Nodes | Out |
tableTwoDimTable | NetworkTestResults | Out |
NodePersistentStorageEntryV3
categorystring | Category name | Out |
namestring | Key name | Out |
sizelong | Size in bytes of value | Out |
timestamp_millislong | Epoch time in milliseconds of when the value was written | Out |
NodePersistentStorageV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
categorystring | Category name | In/Out |
namestring | Key name | In/Out |
valuestring | Value | In/Out |
configuredboolean | Configured | Out |
existsboolean | Exists | Out |
entriesIced[] | List of entries | Out |
NodeV3
h2ostring | IP | Out |
ip_portstring | IP address and port in the form a.b.c.d:e | Out |
healthyboolean | (now-last_ping)<HeartbeatThread.TIMEOUT | Out |
last_pinglong | Time (in msec) of last ping | Out |
pidint | PID | Out |
num_cpusint | num_cpus | Out |
cpus_allowedint | cpus_allowed | Out |
nthreadsint | nthreads | Out |
sys_loadfloat | System load; average #runnables/#cores | Out |
my_cpu_pctint | System CPU percentage used by this H2O process in last interval | Out |
sys_cpu_pctint | System CPU percentage used by everything in last interval | Out |
mem_value_sizelong | Data on Node memory | Out |
pojo_memlong | Temp (non Data) memory | Out |
free_memlong | Free heap | Out |
max_memlong | Maximum memory size for node | Out |
swap_memlong | Size of data on node’s disk | Out |
num_keysint | id="local-keys">local keys< | Out |
free_disklong | Free disk | Out |
max_disklong | Max disk | Out |
rpcs_activeint | Active Remote Procedure Calls | Out |
fjthrdsshort[] | F/J Thread count, by priority | Out |
fjqueueshort[] | F/J Task count, by priority | Out |
tcps_activeint | Open TCP connections | Out |
open_fdsint | Open File Descripters | Out |
gflopsdouble | Linpack GFlops | Out |
mem_bwdouble | Memory Bandwidth | Out |
PCAModelOutputV3
importanceTwoDimTable | Standard deviation and importance of each principal component | In |
eigenvectorsTwoDimTable | Principal components matrix | In |
objectivedouble | Final value of GLRM squared loss function | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
PCAModelV3
model_idKey | Model key | In/Out |
parametersPCAParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputPCAOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
PCAParametersV3
transformenum | Transformation of training data | In |
pca_methodenum | Method for computing PCA (Caution: Power and GLRM are currently experimental and unstable) | In |
kint | Rank of matrix approximation | In/Out |
max_iterationsint | Maximum training iterations | In/Out |
seedlong | RNG seed for initialization | In/Out |
use_all_factor_levelsboolean | Whether first factor level is included in each categorical expansion | In/Out |
compute_metricsboolean | Whether to compute metrics on the training data | In/Out |
impute_missingboolean | Whether to impute missing entries with the column mean | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
PCAV3
parametersPCAParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
ParseSVMLightV3
destination_frameKey | Final frame name | In |
source_framesKey[] | Source frames | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
ParseSetupV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
source_framesKey[] | Source frames | In/Out |
parse_typeenum | Parser type | In/Out |
separatorbyte | Field separator | In/Out |
single_quotesboolean | Single quotes | In/Out |
check_headerint | Check header: 0 means guess, +1 means 1st line is header not data, -1 means 1st line is data not header | In/Out |
column_namesstring[] | Column names | In/Out |
column_typesstring[] | Value types for columns | In/Out |
na_stringsstring[][] | NA strings for columns | In/Out |
column_name_filterstring | Regex for names of columns to return | In/Out |
column_offsetint | Column offset to return | In/Out |
column_countint | Number of columns to return | In/Out |
total_filtered_column_countint | Total number of columns we would return with no column pagination | In/Out |
destination_framestring | Suggested name | Out |
header_lineslong | Number of header lines found | Out |
number_columnsint | Number of columns | Out |
datastring[][] | Sample data | Out |
warningsstring[] | Warnings | Out |
chunk_sizeint | Size of individual parse tasks | Out |
ParseV3
destination_frameKey | Final frame name | In |
source_framesKey[] | Source frames | In |
parse_typeenum | Parser type | In |
separatorbyte | Field separator | In |
single_quotesboolean | Single Quotes | In |
check_headerint | Check header: 0 means guess, +1 means 1st line is header not data, -1 means 1st line is data not header | In |
number_columnsint | Number of columns | In |
column_namesstring[] | Column names | In |
column_typesstring[] | Value types for columns | In |
domainsstring[][] | Domains for categorical columns | In |
na_stringsstring[][] | NA strings for columns | In |
chunk_sizeint | Size of individual parse tasks | In |
delete_on_doneboolean | Delete input key after parse | In |
blockingboolean | Block until the parse completes (as opposed to returning early and requiring polling | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
jobJob | Parse job | Out |
rowslong | Rows | Out |
ProfilerNodeEntryV3
stacktracestring | Stack trace | Out |
countint | Profile Count | Out |
ProfilerNodeV3
node_namestring | Node names | Out |
timestamplong | Timestamp (millis since epoch) | Out |
entriesIced[] | Profile entry list | Out |
ProfilerV3
depthint | Stack trace depth | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
nodesIced[] | (No description available) | Out |
QuantileParametersV3
probsdouble[] | Probabilities for quantiles | In |
combine_methodenum | How to combine quantiles for even sample sizes | In |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
QuantileV3
parametersQuantileParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
RandomDiscreteValueSearchCriteriaV99
seedlong | Seed for random number generator; set to a value other than -1 for reproducibility. | In/Out |
max_modelsint | Maximum number of models to build (optional). | In/Out |
max_runtime_secsdouble | Maximum time to spend building models (optional). | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
strategyenum | Hyperparameter space search strategy. | In/Out |
RapidsFrameV3
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
keyKey | Frame result | Out |
num_rowslong | Rows in Frame result | Out |
num_colsint | Columns in Frame result | Out |
RapidsFunctionV3
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
funstrstring | Function result | Out |
RapidsNumberV3
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
scalardouble | Number result | Out |
RapidsNumbersV3
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
scalardouble[] | Number array result | Out |
RapidsSchema
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
RapidsStringV3
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
stringstring | String result | Out |
RapidsStringsV3
aststring | An Abstract Syntax Tree. | In |
idstring | Key name to assign Frame results | In |
session_idstring | Session key | In |
stringstring[] | String array result | Out |
RapidsV99
aststring | An Abstract Syntax Tree. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
errorstring | Parsing error, if any | Out |
scalardouble | Scalar result | Out |
funstrstring | Function result | Out |
stringstring | String result | Out |
keyKey | Result key | Out |
num_rowslong | Rows in Frame result | Out |
num_colsint | Columns in Frame result | Out |
RemoveAllV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
RemoveV3
keyKey | Object to be removed. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
RequestSchema
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
RouteBase
http_methodstring | (No description available) | Out |
url_patternstring | (No description available) | Out |
summarystring | (No description available) | Out |
handler_classstring | (No description available) | Out |
handler_methodstring | (No description available) | Out |
input_schemastring | (No description available) | Out |
output_schemastring | (No description available) | Out |
doc_methodstring | (No description available) | Out |
path_paramsstring[] | (No description available) | Out |
markdownstring | (No description available) | Out |
RouteV3
http_methodstring | (No description available) | Out |
url_patternstring | (No description available) | Out |
summarystring | (No description available) | Out |
handler_classstring | (No description available) | Out |
handler_methodstring | (No description available) | Out |
input_schemastring | (No description available) | Out |
output_schemastring | (No description available) | Out |
doc_methodstring | (No description available) | Out |
path_paramsstring[] | (No description available) | Out |
markdownstring | (No description available) | Out |
SVDModelOutputV99
v_keyKey | Frame key of right singular vectors | In |
ddouble[] | Singular values | In |
u_keyKey | Frame key of left singular vectors | In |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
SVDModelV99
model_idKey | Model key | In/Out |
parametersSVDParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputSVDOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
SVDParametersV99
transformenum | Transformation of training data | In |
svd_methodenum | Method for computing SVD (Caution: Power and Randomized are currently experimental and unstable) | In |
nvint | Number of right singular vectors | In |
max_iterationsint | Maximum iterations | In |
seedlong | RNG seed for k-means++ initialization | In |
keep_uboolean | Save left singular vectors? | In |
u_namestring | Frame key to save left singular vectors | In |
use_all_factor_levelsboolean | Whether first factor level is included in each categorical expansion | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
SVDV99
parametersSVDParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
Schema
(No fields)
SchemaMetadataBase
versionint | Version number of the Schema. | In |
namestring | Simple name of the Schema. NOTE: the schema_names form a single namespace. | In |
superclassstring | Simple name of the superclass of the Schema. NOTE: the schema_names form a single namespace. | In |
typestring | Simple name of H2O type that this Schema represents. Must not be changed after creation (treat as final). | In |
fieldsFieldMetadata[] | All the public fields of the schema | Out |
markdownstring | Documentation for the schema in Markdown format with GitHub extensions | Out |
SchemaMetadataV3
versionint | Version number of the Schema. | In |
namestring | Simple name of the Schema. NOTE: the schema_names form a single namespace. | In |
superclassstring | Simple name of the superclass of the Schema. NOTE: the schema_names form a single namespace. | In |
typestring | Simple name of H2O type that this Schema represents. Must not be changed after creation (treat as final). | In |
fieldsFieldMetadata[] | All the public fields of the schema | Out |
markdownstring | Documentation for the schema in Markdown format with GitHub extensions | Out |
SharedTreeModelOutputV3
variable_importancesTwoDimTable | Variable Importances | Out |
init_fdouble | The Intercept term, the initial model function value to which trees make adjustments | Out |
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
SharedTreeModelV3
model_idKey | Model key | In/Out |
parametersParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
SharedTreeParametersV3
ntreesint | Number of trees. | In |
max_depthint | Maximum tree depth. | In |
min_rowsdouble | Fewest allowed (weighted) observations in a leaf (in R called ‘nodesize’). | In |
nbinsint | For numerical columns (real/int), build a histogram of (at least) this many bins, then split at the best point | In |
nbins_top_levelint | For numerical columns (real/int), build a histogram of (at most) this many bins at the root level, then decrease by factor of two per level | In |
nbins_catsint | For categorical columns (factors), build a histogram of this many bins, then split at the best point. Higher values can lead to more overfitting. | In |
r2_stoppingdouble | Stop making trees when the R^2 metric equals or exceeds this | In |
seedlong | Seed for pseudo random number generator (if applicable) | In |
build_tree_one_nodeboolean | Run on one node only; no network overhead but fewer cpus used. Suitable for small datasets. | In |
sample_ratedouble | Row sample rate per tree (from 0.0 to 1.0) | In |
sample_rate_per_classdouble[] | Row sample rate per tree per class (from 0.0 to 1.0) | In |
col_sample_rate_per_treedouble | Column sample rate per tree (from 0.0 to 1.0) | In |
col_sample_rate_change_per_leveldouble | Relative change of the column sampling rate for every level (from 0.0 to 2.0) | In |
score_tree_intervalint | Score the model after every so many trees. Disabled if set to 0. | In |
min_split_improvementdouble | Minimum relative improvement in squared error reduction for a split to happen. | In |
random_split_pointsboolean | Whether to use random split points for histograms (to pick the best split from). | In |
balance_classesboolean | Balance training data class counts via over/under-sampling (for imbalanced data). | In/Out |
class_sampling_factorsfloat[] | Desired over/under-sampling ratios per class (in lexicographic order). If not specified, sampling factors will be automatically computed to obtain class balance during training. Requires balance_classes. | In/Out |
max_after_balance_sizefloat | Maximum relative size of the training data after balancing class counts (can be less than 1.0). Requires balance_classes. | In/Out |
max_confusion_matrix_sizeint | Maximum size (# classes) for confusion matrices to be printed in the Logs | In/Out |
max_hit_ratio_kint | Max. number (top K) of predictions to use for hit ratio computation (for multi-class only, 0 to disable) | In/Out |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
SharedTreeV3
parametersParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |
ShutdownV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
SplitFrameV3
keyKey | Job Key | In |
datasetKey | Dataset | In |
ratiosdouble[] | Split ratios - resulting number of split is ratios.length+1 | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
destination_framesKey[] | Destination keys for each output frame split. | In/Out |
StreamingSchema
(No fields)
SynonymV3
keyKey | A word2vec model key. | In |
targetstring | The target string to find synonyms. | In |
cntint | Find the top cnt synonyms of the target word. | In |
synonymsstring[] | The synonyms. | Out |
cos_simfloat[] | The cosine similarities. | Out |
TabulateV3
datasetKey | Dataset | In |
nbins_predictorint | Number of bins for predictor column | In |
nbins_responseint | Number of bins for response column | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
predictorVecSpecifier | Predictor | In/Out |
responseVecSpecifier | Response | In/Out |
weightVecSpecifier | Observation weights (optional) | In/Out |
count_tableTwoDimTable | Counts table | Out |
response_tableTwoDimTable | Response table | Out |
TimelineV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
nowlong | Current time in millis. | Out |
selfstring | This node | Out |
eventsIced[] | recorded timeline events | Out |
TreeStatsV3
min_depthint | minDepth | In |
max_depthint | maxDepth | In |
mean_depthfloat | meanDepth | In |
min_leavesint | minLeaves | In |
max_leavesint | maxLeaves | In |
mean_leavesfloat | meanLeaves | In |
TwoDimTableBase
namestring | Table Name | Out |
descriptionstring | Table Description | Out |
columnsIced[] | Column Specification | Out |
rowcountint | Number of Rows | Out |
dataPolymorphic[][] | Table Data (col-major) | Out |
TwoDimTableV3
namestring | Table Name | Out |
descriptionstring | Table Description | Out |
columnsIced[] | Column Specification | Out |
rowcountint | Number of Rows | Out |
dataPolymorphic[][] | Table Data (col-major) | Out |
TypeaheadV3
srcstring | training_frame | In |
limitint | limit | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
matchesstring[] | matches | Out |
UnlockKeysV3
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
ValidationMessageBase
message_typestring | Type of validation message (ERROR, WARN, INFO, HIDE) | Out |
field_namestring | Field to which the message applies | Out |
messagestring | Message text | Out |
ValidationMessageV3
message_typestring | Type of validation message (ERROR, WARN, INFO, HIDE) | Out |
field_namestring | Field to which the message applies | Out |
messagestring | Message text | Out |
VarImpBase
varimpfloat[] | Variable importance of individual variables | Out |
namesstring[] | Names of variables | Out |
VarImpV3
varimpfloat[] | Variable importance of individual variables | Out |
namesstring[] | Names of variables | Out |
WaterMeterCpuTicksV3
nodeidxint | Index of node to query ticks for (0-based) | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
cpu_tickslong[][] | array of tick counts per core | Out |
WaterMeterIoV3
nodeidxint | Index of node to query ticks for (0-based) | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
persist_statsIced[] | array of IO info | Out |
Word2VecModelOutputV3
namesstring[] | Column names | Out |
domainsstring[][] | Domains for categorical columns | Out |
cross_validation_modelsKey | Cross-validation models (model ids) | Out |
cross_validation_predictionsKey[] | Cross-validation predictions, one per cv model (deprecated, use cross_validation_holdout_predictions_frame_id instead) | Out |
cross_validation_holdout_predictions_frame_idKey | Cross-validation holdout predictions (full out-of-sample predictions on training data) | Out |
cross_validation_fold_assignment_frame_idKey | Cross-validation fold assignment (each row is assigned to one holdout fold) | Out |
model_categoryenum | Category of the model (e.g., Binomial) | Out |
model_summaryTwoDimTable | Model summary | Out |
scoring_historyTwoDimTable | Scoring history | Out |
training_metricsModelMetrics | Training data model metrics | Out |
validation_metricsModelMetrics | Validation data model metrics | Out |
cross_validation_metricsModelMetrics | Cross-validation model metrics | Out |
cross_validation_metrics_summaryTwoDimTable | Cross-validation model metrics summary | Out |
statusstring | Job status | Out |
start_timelong | Start time in milliseconds | Out |
end_timelong | End time in milliseconds | Out |
run_timelong | Runtime in milliseconds | Out |
helpMap | Help information for output fields | Out |
Word2VecModelV3
model_idKey | Model key | In/Out |
parametersWord2VecParameters | The build parameters for the model (e.g. K for KMeans). | Out |
outputWord2VecOutput | The build output for the model (e.g. the cluster centers for KMeans). | Out |
compatible_framesstring[] | Compatible frames, if requested | Out |
checksumlong | Checksum for all the things that go into building the Model. | Out |
algostring | The algo name for this Model. | Out |
algo_full_namestring | The pretty algo name for this Model (e.g., Generalized Linear Model, rather than GLM). | Out |
response_column_namestring | The response column name for this Model (if applicable). Is null otherwise. | Out |
data_frameKey | The Model’s training frame key | Out |
timestamplong | Timestamp for when this model was completed | Out |
Word2VecParametersV3
vecSizeint | Set size of word vectors | In |
windowSizeint | Set max skip length between words | In |
sentSampleRatefloat | Set threshold for occurrence of words. Those that appear with higher frequency in the training data will be randomly down-sampled; useful range is (0, 1e-5) | In |
normModelenum | Use Hierarchical Softmax or Negative Sampling | In |
negSampleCntint | Number of negative examples, common values are 3 - 10 (0 = not used) | In |
epochsint | Number of training iterations to run | In |
minWordFreqint | This will discard words that appear less than | In |
initLearningRatefloat | Set the starting learning rate | In |
wordModelenum | Use the continuous bag of words model or the Skip-Gram model | In |
model_idKey | Destination id for this model; auto-generated if not specified | In/Out |
training_frameKey | Training frame | In/Out |
validation_frameKey | Validation frame | In/Out |
nfoldsint | Number of folds for N-fold cross-validation | In/Out |
keep_cross_validation_predictionsboolean | Keep cross-validation model predictions | In/Out |
keep_cross_validation_fold_assignmentboolean | Keep cross-validation fold assignment | In/Out |
parallelize_cross_validationboolean | Allow parallel training of cross-validation models | In/Out |
response_columnVecSpecifier | Response column | In/Out |
weights_columnVecSpecifier | Column with observation weights | In/Out |
offset_columnVecSpecifier | Offset column | In/Out |
fold_columnVecSpecifier | Column with cross-validation fold index assignment per observation | In/Out |
fold_assignmentenum | Cross-validation fold assignment scheme, if fold_column is not specified | In/Out |
ignored_columnsstring[] | Ignored columns | In/Out |
ignore_const_colsboolean | Ignore constant columns | In/Out |
score_each_iterationboolean | Whether to score during each iteration of model training | In/Out |
checkpointKey | Model checkpoint to resume training with | In/Out |
stopping_roundsint | Early stopping based on convergence of stopping_metric. Stop if simple moving average of length k of the stopping_metric does not improve for k:=stopping_rounds scoring events (0 to disable) | In/Out |
max_runtime_secsdouble | Maximum allowed runtime in seconds for model training. Use 0 to disable. | In/Out |
stopping_metricenum | Metric to use for early stopping (AUTO: logloss for classification, deviance for regression) | In/Out |
stopping_tolerancedouble | Relative tolerance for metric-based stopping criterion (stop if relative improvement is not at least this much) | In/Out |
Word2VecV3
parametersWord2VecParameters | Model builder parameters. | In |
_exclude_fieldsstring | Comma-separated list of JSON field paths to exclude from the result, used like: “/3/Frames?_exclude_fields=frames/frame_id/URL,__meta” | In |
algostring | The algo name for this ModelBuilder. | Out |
algo_full_namestring | The pretty algo name for this ModelBuilder (e.g., Generalized Linear Model, rather than GLM). | Out |
can_buildenum[] | Model categories this ModelBuilder can build. | Out |
visibilityenum | Should the builder always be visible, be marked as beta, or only visible if the user starts up with the experimental flag? | Out |
jobJob | Job Key | Out |
messagesValidationMessage[] | Parameter validation messages | Out |
error_countint | Count of parameter validation errors | Out |
__http_statusint | HTTP status to return for this build. | Out |

