FAQ¶
General Troubleshooting Tips¶
Confirm your internet connection is active.
Test connectivity using curl: First, log in to the first node and enter
curl http://<Node2IP>:54321(where<Node2IP>is the IP address of the second node. Then, log in to the second node and entercurl http://<Node1IP>:54321(where<Node1IP>is the IP address of the first node). Look for output from H2O.Try allocating more memory to H2O by modifying the
-Xmxvalue when launching H2O from the command line (for example,java -Xmx10g -jar h2o.jarallocates 10g of memory for H2O). If you create a cluster with four 20g nodes (by specifying-Xmx20gfour times), H2O will have a total of 80 gigs of memory available. For best performance, we recommend sizing your cluster to be about four times the size of your data. To avoid swapping, the-Xmxallocation must not exceed the physical memory on any node. Allocating the same amount of memory for all nodes is strongly recommended, as H2O works best with symmetric nodes.Confirm that no other sessions of H2O are running. To stop all running H2O sessions, enter
ps -efww | grep h2oin your shell (OSX or Linux).Confirm ports 54321 and 54322 are available for both TCP and UDP. Launch Telnet (for Windows users) or Terminal (for OS X users), then type
telnet localhost 54321,telnet localhost 54322Confirm your firewall is not preventing the nodes from locating each other. If you can’t launch H2O, we recommend temporarily disabling any firewalls until you can confirm they are not preventing H2O from launching.
Confirm the nodes are not using different versions of H2O. If the H2O initialization is not successful, look at the output in the shell - if you see
Attempting to join /localhost:54321 with an H2O version mismatch (md5 differs), update H2O on all the nodes to the same version.Confirm that there is space in the
/tmpdirectory.- Windows: In Command Prompt, enter
TEMPand%TEMP%and delete files as needed, or use Disk Cleanup. - OS X: In Terminal, enter
open $TMPDIRand delete the folder with your username.
- Windows: In Command Prompt, enter
Confirm that the username is the same on all nodes; if not, define the cloud in the terminal when launching using
-name:java -jar h2o.jar -name myCloud.Confirm that there are no spaces in the file path name used to launch H2O.
Confirm that the nodes are not on different networks by confirming that the IP addresses of the nodes are the same in the output:
INFO: Listening for HTTP and REST traffic on IP_Address/ 06-18 10:54:21.586 192.168.1.70:54323 25638 main INFO: H2O cloud name: 'H2O_User' on IP_Address, discovery address /Discovery_Address INFO: Cloud of size 1 formed [IP_Address]
Check if the nodes have different interfaces; if so, use the -network option to define the network (for example,
-network 127.0.0.1). To use a network range, use a comma to separate the IP addresses (for example,-network 123.45.67.0/22,123.45.68.0/24).Force the bind address using
-ip:java -jar h2o.jar -ip <IP_Address> -port <PortNumber>.(Hadoop only) Try launching H2O with a longer timeout:
hadoop jar h2odriver.jar -timeout 1800(Hadoop only) Try to launch H2O using more memory:
hadoop jar h2odriver.jar -mapperXmx 10g. The cluster’s memory capacity is the sum of all H2O nodes in the cluster.(Linux only) Check if you have SELINUX or IPTABLES enabled; if so, disable them.
(EC2 only) Check the configuration for the EC2 security group.
The following error message displayed when I tried to launch H2O - what should I do?
Exception in thread "main" java.lang.UnsupportedClassVersionError: water/H2OApp
: Unsupported major.minor version 51.0
at java.lang.ClassLoader.defineClass1(Native Method)
at java.lang.ClassLoader.defineClassCond(Unknown Source)
at java.lang.ClassLoader.defineClass(Unknown Source)
at java.security.SecureClassLoader.defineClass(Unknown Source)
at java.net.URLClassLoader.defineClass(Unknown Source)
at java.net.URLClassLoader.access$000(Unknown Source)
at java.net.URLClassLoader$1.run(Unknown Source)
at java.security.AccessController.doPrivileged(Native Method)
at java.net.URLClassLoader.findClass(Unknown Source)
at java.lang.ClassLoader.loadClass(Unknown Source)
at sun.misc.Launcher$AppClassLoader.loadClass(Unknown Source)
at java.lang.ClassLoader.loadClass(Unknown Source)
Could not find the main class: water.H2OApp. Program will exit.
This error output indicates that your Java version is not supported. Upgrade to Java 7 (JVM) or later and H2O should launch successfully.
I am not launching on Hadoop. How can i increase the amount of time that H2O allows for expected nodes to connect?
For cluster startup, if you are not launching on Hadoop, then you will not need to specify a timeout. You can add additional nodes to the cloud as long as you haven’t submitted any jobs to the cluster. When you do submit a job to the cluster, the cluster will lock and will print a message similar to “Locking cloud to new members, because <reason>...”.
What’s the best approach to help diagnose a possible memory problem on a cluster?
We’ve found that the best way to understand JVM memory consumption is to turn on the following JVM flags:
-verbose:gc -XX:+PrintGCDetails -XX:+PrintGCTimeStamps
You can then use the following tool to analyze the output: http://www.tagtraum.com/gcviewer-download.html
How can I debug memory issues?
We recommend the following approach using R to debug memory issues:
my for loop {
# perform loop
rm(R object that isn’t needed anymore)
rm(R object of h2o thing that isn’t needed anymore)
# trigger removal of h2o back-end objects that got rm’d above, since the rm can be lazy.
gc()
# optional extra one to be paranoid. this is usually very fast.
gc()
# optionally sanity check that you see only what you expect to see here, and not more.
h2o.ls()
# tell back-end cluster nodes to do three back-to-back JVM full GCs.
h2o:::.h2o.garbageCollect()
h2o:::.h2o.garbageCollect()
h2o:::.h2o.garbageCollect()
}
Note that the h2o.garbageCollct() function works as follows:
# Trigger an explicit garbage collection across all nodes in the H2O cluster.
.h2o.garbageCollect <- function() {
res <- .h2o.__remoteSend("GarbageCollect", method = "POST")
}
This tells the backend to do a forcible full-GC on each node in the H2O cluster. Doing three of them back-to-back makes it stand out clearly in the gcviewer chart where the bottom-of-inner loop is. You can then correlate what you expect to see with the X (time) axis of the memory utilization graph.
At this point you want to see if the bottom trough of the usage is growing from iteration to iteration after the triple full-GC bars in the graph. If the trough is not growing from iteration to iteration, then there is no leak; your usage is just really too much, and you need a bigger heap. If the trough is growing, then there is likely some kind of leak. You can try to use h2o.ls() to learn where the leak is. If h2o.ls() doesn’t help, then you will have to drill much deeper using, for example, YourKit and reviewing the JVM-level heap profiles.
Algorithms¶
What’s the process for implementing new algorithms in H2O?
This blog post by Cliff walks you through building a new algorithm, using K-Means, Quantiles, and Grep as examples.
To learn more about performance characteristics when implementing new algorithms, refer to Cliff’s KV Store Guide.
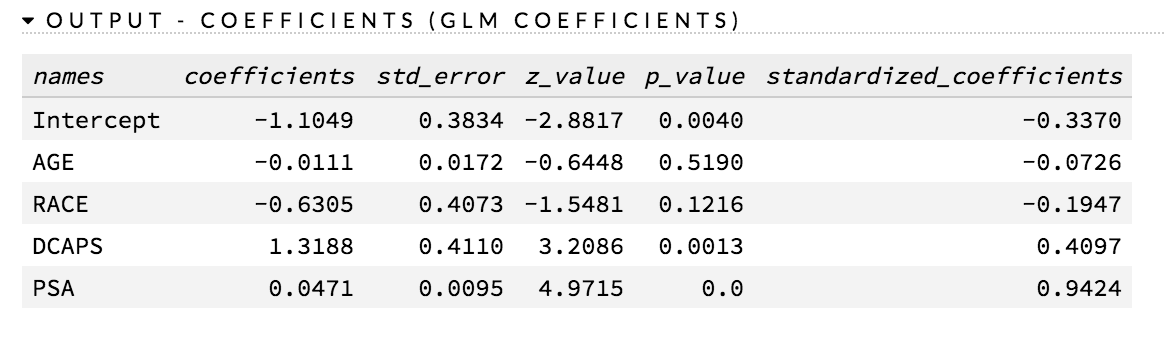
How do I find the standard errors of the parameter estimates (p-values)?
P-values are currently supported for non-regularized GLM. The following requirements must be met:
- The family cannot be multinomial
- The lambda value must be equal to zero
- The IRLSM solver must be used
- Lambda search cannot be used
To generate p-values, do one of the following:
- check the compute_p_values checkbox in the GLM model builder in Flow
- use
compute_p_values=TRUEin R or Python while creating the model
The p-values are listed in the coefficients table (as shown in the following example screenshot):
How do I specify regression or classification for Distributed Random Forest in the web UI?
If the response column is numeric, H2O generates a regression model. If the response column is enum, the model uses classification. To specify the column type, select it from the drop-down column name list in the Edit Column Names and Types section during parsing.
What’s the largest number of classes that H2O supports for multinomial prediction?
For tree-based algorithms, the maximum number of classes (or levels) for a response column is 1000.
How do I obtain a tree diagram of my DRF model?
Output the SVG code for the edges and nodes. A simple tree visitor is available here and the Java code generator is available here.
Is Word2Vec available? I can see the Java and R sources, but calling the API generates an error.
Word2Vec, along with other natural language processing (NLP) algos, are currently in development in the current version of H2O.
What are the “best practices” for preparing data for a K-Means model?
There aren’t specific “best practices,” as it depends on your data and the column types. However, removing outliers and transforming any categorical columns to have the same weight as the numeric columns will help, especially if you’re standardizing your data.
What is your implementation of Deep Learning based on?
Our Deep Learning algorithm is based on the feedforward neural net. For more information, refer to our Data Science documentation or Wikipedia.
How is deviance computed for a Deep Learning regression model?
For a Deep Learning regression model, deviance is computed as follows:
Loss = MeanSquare -> MSE==Deviance For Absolute/Laplace or Huber -> MSE != Deviance.
For my 0-tree GBM multinomial model, I got a different score depending on whether or not validation was enabled, even though my dataset was the same - why is that?
Different results may be generated because of the way H2O computes the initial MSE.
How does your Deep Learning Autoencoder work? Is it deep or shallow?
H2O’s DL autoencoder is based on the standard deep (multi-layer) neural net architecture, where the entire network is learned together, instead of being stacked layer-by-layer. The only difference is that no response is required in the input and that the output layer has as many neurons as the input layer. If you don’t achieve convergence, then try using the Tanh activation and fewer layers. We have some example test scripts here, and even some that show how stacked auto-encoders can be implemented in R.
Are there any H2O examples using text for classification?
Currently, the following examples are available for Sparkling Water:
- Use TF-IDF weighting scheme for classifying text messages
https://github.com/h2oai/sparkling-water/blob/master/examples/scripts/hamOrSpam.script.scala
- Use Word2Vec Skip-gram model + GBM for classifying job titles
Most machine learning tools cannot predict with a new categorical level that was not included in the training set. How does H2O make predictions in this scenario?
Here is an example of how the prediction process works in H2O:
- Train a model using data that has a categorical predictor column with levels B,C, and D (no other levels); this level will be the “training set domain”: {B,C,D}
- During scoring, the test set has only rows with levels A,C, and E for that column; this is the “test set domain”: {A,C,E}
- For scoring, a combined “scoring domain” is created, which is the training domain appended with the extra test set domain entries: {B,C,D,A,E}
- Each model can handle these extra levels {A,E} separately during scoring.
The behavior for unseen categorical levels depends on the algorithm and how it handles missing levels (NA values):
- For DRF and GBM, missing values are interpreted as containing information (i.e., missing for a reason) rather than missing at random. During tree building, split decisions for every node are found by minimizing the loss function and treating missing values as a separate category that can go either left or right.
- Deep Learning creates an extra input neuron for missing and unseen categorical levels, which can remain untrained if there were no missing or unseen categorical levels in the training data, resulting in a random contribution to the next layer during testing.
- GLM skips unseen levels in the beta*x dot product.
How are quantiles computed?
The quantile results in Flow are computed lazily on-demand and cached.
It is a fast approximation (max - min / 1024) that is very accurate for
most use cases. If the distribution is skewed, the quantile results may
not be as accurate as the results obtained using h2o.quantile in R
or H2OFrame.quantile in Python.
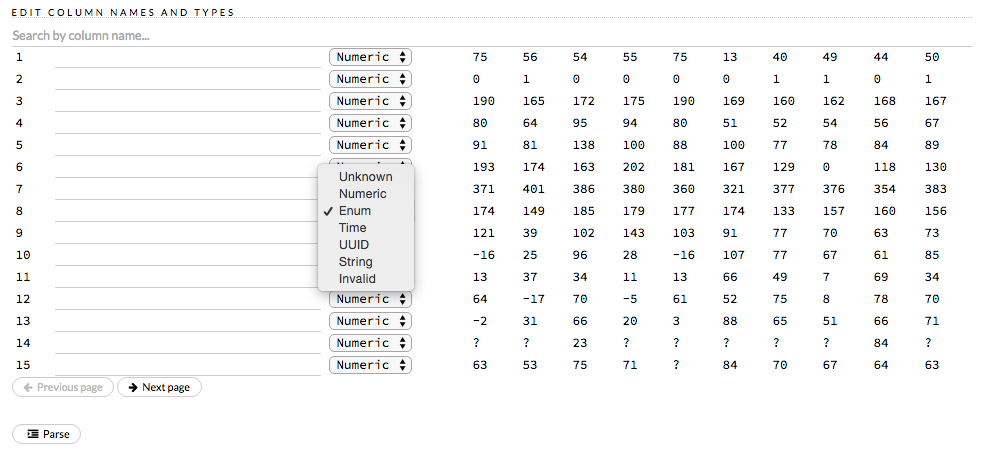
How do I create a classification model? The model always defaults to regression.
To create a classification model, the response column type must be
enum - if the response is numeric, a regression model is
created.
To convert the response column:
- Before parsing, click the drop-down menu to the right of the column
name or number and select
Enum
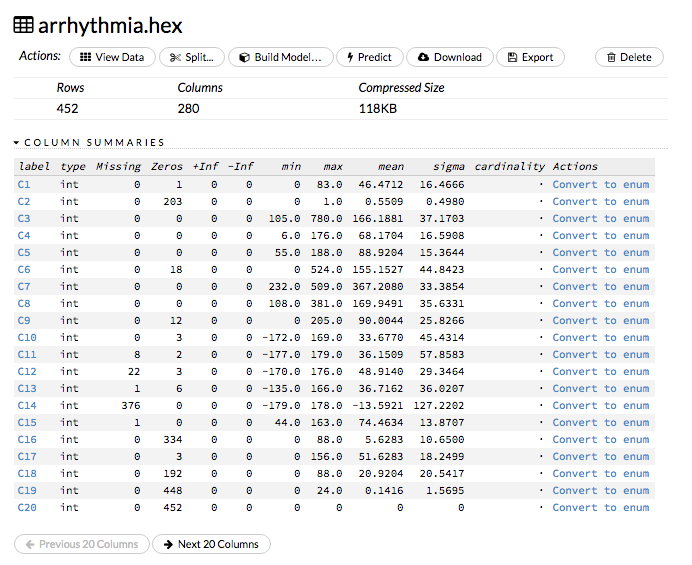
or
- Click on the .hex link for the data frame (or use the
getFrameSummary "<frame_name>.hex"command, where<frame_name>is the name of the frame), then click the Convert to enum link to the right of the column name or number
Building H2O¶
During the build process, the following error message displays. What do I need to do to resolve it?
Error: Missing name at classes.R:19
In addition: Warning messages:
1: @S3method is deprecated. Please use @export instead
2: @S3method is deprecated. Please use @export instead
Execution halted
To build H2O, Roxygen2 version 4.1.1 is required.
To update your Roxygen2 version, install the versions package in R,
then use install.versions("roxygen2", "4.1.1").
Using ``./gradlew build`` doesn’t generate a build successfully - is there anything I can do to troubleshoot?
Use ./gradlew clean before running ./gradlew build.
I tried using ``./gradlew build`` after using ``git pull`` to update my local H2O repo, but now I can’t get H2O to build successfully - what should I do?
Try using ./gradlew build -x test - the build may be failing tests
if data is not synced correctly.
Clusters¶
When trying to launch H2O, I received the following error message: ``ERROR: Too many retries starting cloud.`` What should I do?
If you are trying to start a multi-node cluster where the nodes use multiple network interfaces, by default H2O will resort to using the default host (127.0.0.1).
To specify an IP address, launch H2O using the following command:
java -jar h2o.jar -ip <IP_Address> -port <PortNumber>
If this does not resolve the issue, try the following additional troubleshooting tips:
- Confirm your internet connection is active.
- Test connectivity using curl: First, log in to the first node and enter curl http://:54321 (where is the IP address of the second node. Then, log in to the second node and enter curl http://:54321 (where is the IP address of the first node). Look for output from H2O.
- Confirm ports 54321 and 54322 are available for both TCP and UDP.
- Confirm your firewall is not preventing the nodes from locating each other.
- Confirm the nodes are not using different versions of H2O.
- Confirm that the username is the same on all nodes; if not, define
the cloud in the terminal when launching using
-name:java -jar h2o.jar -name myCloud. - Confirm that the nodes are not on different networks.
- Check if the nodes have different interfaces; if so, use the -network
option to define the network (for example,
-network 127.0.0.1). - Force the bind address using
-ip:java -jar h2o.jar -ip <IP_Address> -port <PortNumber>. - (Linux only) Check if you have SELINUX or IPTABLES enabled; if so, disable them.
- (EC2 only) Check the configuration for the EC2 security group.
What should I do if I tried to start a cluster but the nodes started independent clouds that are not connected?
Because the default cloud name is the user name of the node, if the
nodes are on different operating systems (for example, one node is using
Windows and the other uses OS X), the different user names on each
machine will prevent the nodes from recognizing that they belong to the
same cloud. To resolve this issue, use -name to configure the same
name for all nodes.
One of the nodes in my cluster is unavailable — what do I do?
H2O does not support high availability (HA). If a node in the cluster is unavailable, bring the cluster down and create a new healthy cluster.
How do I add new nodes to an existing cluster?
New nodes can only be added if H2O has not started any jobs. Once H2O starts a task, it locks the cluster to prevent new nodes from joining. If H2O has started a job, you must create a new cluster to include additional nodes.
How do I check if all the nodes in the cluster are healthy and communicating?
In the Flow web UI, click the Admin menu and select Cluster Status.
How do I create a cluster behind a firewall?
H2O uses two ports:
- The
REST_APIport (54321): Specify when launching H2O using-port; uses TCP only. - The
INTERNAL_COMMUNICATIONport (54322): Implied based on the port specified as theREST_APIport, +1; requires TCP and UDP.
You can start the cluster behind the firewall, but to reach it, you must
make a tunnel to reach the REST_API port. To use the cluster, the
REST_API port of at least one node must be reachable.
How can I create a multi-node H2O cluster on a SLURM system?
The syntax below comes from https://github.com/ck37/savio-notes/blob/master/h2o-slurm-multinode.Rmd and describes how to create a multi-node H2O cluster on a Simple Linux Utility for Resource Management (SLURM) system using R.
# Check on the nodes we have access to.
node_list = Sys.getenv("SLURM_NODELIST")
cat("SLURM nodes:", node_list, "\n")
# Loop up IPs of the allocated nodes.
if (node_list != "") {
nodes = strsplit(node_list, ",")[[1]]
ips = rep(NA, length(nodes))
for (i in 1:length(nodes)) {
args = c("hosts", nodes[i])
result = system2("getent", args = args, stdout = T)
# Extract the IP from the result output.
ips[i] = sub("^([^ ]+) +.*$", "\\1", result, perl = T)
}
cat("SLURM IPs:", paste(ips, collapse=", "), "\n")
# Combine into a network string for h2o.
network = paste0(paste0(ips, "/32"), collapse=",")
cat("Network:", network, "\n")
}
# Specify how many nodes we want h2o to use.
h2o_num_nodes = length(ips)
# Options to pass to java call:
args = c(
# -Xmx30g allocate 30GB of RAM per node. Needs to come before "-jar"
"-Xmx30g",
# Specify path to downloaded h2o jar.
"-jar ~/software/h2o-latest/h2o.jar",
# Specify a cloud name for the cluster.
"-name h2o_r",
# Specify IPs of other nodes.
paste("-network", network)
)
cat(paste0("Args:\n", paste(args, collapse="\n"), "\n"))
# Run once for each node we want to start.
for (node_i in 1:h2o_num_nodes) {
cat("\nLaunching h2o worker on", ips[node_i], "\n")
new_args = c(ips[node_i], "java", args)
# Ssh into the target IP and launch an h2o worker with its own
# output and error files. These could go in a subdirectory.
cmd_result = system2("ssh", args = new_args,
stdout = paste0("h2o_out_", node_i, ".txt"),
stderr = paste0("h2o_err_", node_i, ".txt"),
# Need to specify wait=F so that it runs in the background.
wait = F)
# This should be 0.
cat("Cmd result:", cmd_result, "\n")
# Wait one second between inits.
Sys.sleep(1L)
}
# Wait 3 more seconds to find all the nodes, otherwise we may only
# find the node on localhost.
Sys.sleep(3L)
# Check if h2o is running. We will see ssh processes and one java process.
system2("ps", c("-ef", "| grep h2o.jar"), stdout = T)
suppressMessages(library(h2oEnsemble))
# Connect to our existing h2o cluster.
# Do not try to start a new server from R.
h2o.init(startH2O = F)
#################################
# Run H2O commands here.
#################################
h2o.shutdown(prompt = F)
I launched H2O instances on my nodes - why won’t they form a cloud?
If you launch without specifying the IP address by adding argument -ip:
$ java -Xmx20g -jar h2o.jar -flatfile flatfile.txt -port 54321
and multiple local IP addresses are detected, H2O uses the default localhost (127.0.0.1) as shown below:
10:26:32.266 main WARN WATER: Multiple local IPs detected:
+ /198.168.1.161 /198.168.58.102
+ Attempting to determine correct address...
10:26:32.284 main WARN WATER: Failed to determine IP, falling back to localhost.
10:26:32.325 main INFO WATER: Internal communication uses port: 54322
+ Listening for HTTP and REST traffic
+ on http://127.0.0.1:54321/
10:26:32.378 main WARN WATER: Flatfile configuration does not include self:
/127.0.0.1:54321 but contains [/192.168.1.161:54321, /192.168.1.162:54321]
To avoid using 127.0.0.1 on servers with multiple local IP addresses, run the command with the -ip argument to force H2O to launch at the specified IP:
$ java -Xmx20g -jar h2o.jar -flatfile flatfile.txt -ip 192.168.1.161 -port 54321
How does the timeline tool work?
The timeline is a debugging tool that provides information on the current communication between H2O nodes. It shows a snapshot of the most recent messages passed between the nodes. Each node retains its own history of messages sent to or received from other nodes.
H2O collects these messages from all the nodes and orders them by whether they were sent or received. Each node has an implicit internal order where sent messages must precede received messages on the other node.
The following information displays for each message:
HH:MM:SS:MSandnanosec: The local time of the eventWho: The endpoint of the message; can be either a source/receiver node or source node and multicast for broadcasted messagesI/O Type: The type of communication (either UDP for small messages or TCP for large messages) >Note: UDP messages are only sent if the UDP option was enabled when launching H2O or for multicast when a flatfile is not used for configuration.Event: The type of H2O message. The most common type is a distributed task, which displays asexec(the requested task) ->ack(results of the processed task) ->ackck(sender acknowledges receiving the response, task is completed and removed)rebooted: Sent during node startupheartbeat: Provides small message tracking information about node health, exchanged periodically between nodesfetchack: Aknowledgement of theFetchtype task, which retrieves the ID of a previously unseen typebytes: Information extracted from the message, including the type of the task and the unique task number
Data¶
How should I format my SVMLight data before importing?
The data must be formatted as a sorted list of unique integers, the column indices must be >= 1, and the columns must be in ascending order.
Does H2O provide native sparse support?
Sparse data is supported natively by loading a sparse matrix from an SVMLight file. In addition, H2O includes a direct conversion of a sparse matrix to an H2O Frame in Python via the h2o.H2OFrame() method and in R via the as.h2o() function. For sparse data, H2O writes a sparse matrix to SVMLight format and then loads it back in using h2o.import_file in Python or h2o.importFile with parse_type=SVMLight in R.
In R, a sparse matrix is specified using Matrix::sparseMatrix along with the boolean flag, sparse=TRUE. For example:
data <- rep(0, 100) data[(1:10)^2] <- 1:10 * pi m <- matrix(data, ncol = 20, byrow = TRUE) m <- Matrix::Matrix(m, sparse = TRUE) h2o.matrix <- as.h2o(m, "sparse_matrix")
In Python, a sparse matrix is specified using scipy.parse. For example:
import scipy.sparse as sp A = sp.csr_matrix([[1, 2, 0, 5.5], [0, 0, 3, 6.7], [4, 0, 5, 0]]) fr = h2o.H2OFrame(A) A = sp.lil_matrix((1000, 1000)) A.setdiag(10) for i in range(999): A[i, i + 1] = -3 A[i + 1, i] = -2 fr = h2o.H2OFrame(A)
What date and time formats does H2O support?
H2O is set to auto-detect two major date/time formats. Because many date time formats are ambiguous (e.g. 01/02/03), general date time detection is not used.
The first format is for dates formatted as yyyy-MM-dd. Year is a four-digit number, the month is a two-digit number ranging from 1 to 12, and the day is a two-digit value ranging from 1 to 31. This format can also be followed by a space and then a time (specified below).
The second date format is for dates formatted as dd-MMM-yy. Here the day must be one or two digits with a value ranging from 1 to 31. The month must be either a three-letter abbreviation or the full month name but is not case sensitive. The year must be either two or four digits. In agreement with POSIX standards, two-digit dates >= 69 are assumed to be in the 20th century (e.g. 1969) and the rest are part of the 21st century. This date format can be followed by either a space or colon character and then a time. The ‘-‘ between the values is optional.
Times are specified as HH:mm:ss. HH is a two-digit hour and must be a value between 0-23 (for 24-hour time) or 1-12 (for a twelve-hour clock). mm is a two-digit minute value and must be a value between 0-59. ss is a two-digit second value and must be a value between 0-59. This format can be followed with either milliseconds, nanoseconds, and/or the cycle (i.e. AM/PM). If milliseconds are included, the format is HH:mm:ss:SSS. If nanoseconds are included, the format is HH:mm:ss:SSSnnnnnn. H2O only stores fractions of a second up to the millisecond, so accuracy may be lost. Nanosecond parsing is only included for convenience. Finally, a valid time can end with a space character and then either “AM” or “PM”. For this format, the hours must range from 1 to 12. Within the time, the ‘:’ character can be replaced with a ‘.’ character.
How does H2O handle name collisions/conflicts in the dataset?
If there is a name conflict (for example, column 48 isn’t named, but C48 already exists), then the column name in concatenated to itself until a unique name is created. So for the previously cited example, H2O will try renaming the column to C48C48, then C48C48C48, and so on until an unused name is generated.
What types of data columns does H2O support?
Currently, H2O supports:
- float (any IEEE double)
- integer (up to 64bit, but compressed according to actual range)
- factor (same as integer, but with a String mapping, often handled differently in the algorithms)
- time (same as 64bit integer, but with a time-since-Unix-epoch interpretation)
- UUID (128bit integer, no math allowed)
- String
I am trying to parse a Gzip data file containing multiple files, but it does not parse as quickly as the uncompressed files. Why is this?
Parsing Gzip files is not done in parallel, so it is sequential and uses only one core. Other parallel parse compression schemes are on the roadmap.
General¶
How do I score using an exported JSON model?
Since JSON is just a representation format, it cannot be directly executed, so a JSON export can’t be used for scoring. However, you can score by:
- including the POJO in your execution stream and handing it observations one at a time
or
- handing your data in bulk to an H2O cluster, which will score using high throughput parallel and distributed bulk scoring.
How do I score using an exported POJO?
The generated POJO can be used indepedently of a H2O cluster. First use
curl to send the h2o-genmodel.jar file and the java code for model
to the server. The following is an example; the ip address and model
names will need to be changed.
mkdir tmpdir
cd tmpdir
curl http://127.0.0.1:54321/3/h2o-genmodel.jar > h2o-genmodel.jar
curl http://127.0.0.1:54321/3/Models.java/gbm_model > gbm_model.java
To score a simple .CSV file, download the PredictCsv.java file and compile it with the POJO. Make a subdirectory for the compilation (this is useful if you have multiple models to score on).
wget https://raw.githubusercontent.com/h2oai/h2o-3/master/h2o-r/tests/testdir_javapredict/PredictCSV.java
mkdir gbm_model_dir
javac -cp h2o-genmodel.jar -J-Xmx2g -J-XX:MaxPermSize=128m PredictCSV.java gbm_model.java -d gbm_model_dir
Specify the following: - the classpath using -cp - the model name
(or class) using --model - the csv file you want to score using
--input - the location for the predictions using --output.
You must match the table column names to the order specified in the POJO. The output file will be in a .hex format, which is a lossless text representation of floating point numbers. Both R and Java will be able to read the hex strings as numerics.
java -ea -cp h2o-genmodel.jar:gbm_model_dir -Xmx4g -XX:MaxPermSize=256m -XX:ReservedCodeCacheSize=256m PredictCSV --header --model gbm_model --input input.csv --output output.csv
How do I predict using multiple response variables?
Currently, H2O does not support multiple response variables. To predict different response variables, build multiple models.
How do I kill any running instances of H2O?
In Terminal, enter ps -efww | grep h2o, then kill any running PIDs.
You can also find the running instance in Terminal and press Ctrl +
C on your keyboard. To confirm no H2O sessions are still running, go
to http://localhost:54321 and verify that the H2O web UI does not
display.
Why is H2O not launching from the command line?
$ java -jar h2o.jar &
% Exception in thread "main" java.lang.ExceptionInInitializerError
at java.lang.Class.initializeClass(libgcj.so.10)
at water.Boot.getMD5(Boot.java:73)
at water.Boot.<init>(Boot.java:114)
at water.Boot.<clinit>(Boot.java:57)
at java.lang.Class.initializeClass(libgcj.so.10)
Caused by: java.lang.IllegalArgumentException
at java.util.regex.Pattern.compile(libgcj.so.10)
at water.util.Utils.<clinit>(Utils.java:1286)
at java.lang.Class.initializeClass(libgcj.so.10)
...4 more
The only prerequisite for running H2O is a compatible version of Java. We recommend Oracle’s Java 1.7.
Why did I receive the following error when I tried to launch H2O?
[root@sandbox h2o-dev-0.3.0.1188-hdp2.2]hadoop jar h2odriver.jar -nodes 2 -mapperXmx 1g -output hdfsOutputDirName
Determining driver host interface for mapper->driver callback...
[Possible callback IP address: 10.0.2.15]
[Possible callback IP address: 127.0.0.1]
Using mapper->driver callback IP address and port: 10.0.2.15:41188
(You can override these with -driverif and -driverport.)
Memory Settings:
mapreduce.map.java.opts: -Xms1g -Xmx1g -Dlog4j.defaultInitOverride=true
Extra memory percent: 10
mapreduce.map.memory.mb: 1126
15/05/08 02:33:40 INFO impl.TimelineClientImpl: Timeline service address: http://sandbox.hortonworks.com:8188/ws/v1/timeline/
15/05/08 02:33:41 INFO client.RMProxy: Connecting to ResourceManager at sandbox.hortonworks.com/10.0.2.15:8050
15/05/08 02:33:47 INFO mapreduce.JobSubmitter: number of splits:2
15/05/08 02:33:48 INFO mapreduce.JobSubmitter: Submitting tokens for job: job_1431052132967_0001
15/05/08 02:33:51 INFO impl.YarnClientImpl: Submitted application application_1431052132967_0001
15/05/08 02:33:51 INFO mapreduce.Job: The url to track the job: http://sandbox.hortonworks.com:8088/proxy/application_1431052132967_0001/
Job name 'H2O_3889' submitted
JobTracker job ID is 'job_1431052132967_0001'
For YARN users, logs command is 'yarn logs -applicationId application_1431052132967_0001'
Waiting for H2O cluster to come up...
H2O node 10.0.2.15:54321 requested flatfile
ERROR: Timed out waiting for H2O cluster to come up (120 seconds)
ERROR: (Try specifying the -timeout option to increase the waiting time limit)
15/05/08 02:35:59 INFO impl.TimelineClientImpl: Timeline service address: http://sandbox.hortonworks.com:8188/ws/v1/timeline/
15/05/08 02:35:59 INFO client.RMProxy: Connecting to ResourceManager at sandbox.hortonworks.com/10.0.2.15:8050
----- YARN cluster metrics -----
Number of YARN worker nodes: 1
----- Nodes -----
Node: http://sandbox.hortonworks.com:8042 Rack: /default-rack, RUNNING, 1 containers used, 0.2 / 2.2 GB used, 1 / 8 vcores used
----- Queues -----
Queue name: default
Queue state: RUNNING
Current capacity: 0.11
Capacity: 1.00
Maximum capacity: 1.00
Application count: 1
----- Applications in this queue -----
Application ID: application_1431052132967_0001 (H2O_3889)
Started: root (Fri May 08 02:33:50 UTC 2015)
Application state: FINISHED
Tracking URL: http://sandbox.hortonworks.com:8088/proxy/application_1431052132967_0001/jobhistory/job/job_1431052132967_0001
Queue name: default
Used/Reserved containers: 1 / 0
Needed/Used/Reserved memory: 0.2 GB / 0.2 GB / 0.0 GB
Needed/Used/Reserved vcores: 1 / 1 / 0
Queue 'default' approximate utilization: 0.2 / 2.2 GB used, 1 / 8 vcores used
----------------------------------------------------------------------
ERROR: Job memory request (2.2 GB) exceeds available YARN cluster memory (2.2 GB)
WARNING: Job memory request (2.2 GB) exceeds queue available memory capacity (2.0 GB)
ERROR: Only 1 out of the requested 2 worker containers were started due to YARN cluster resource limitations
----------------------------------------------------------------------
Attempting to clean up hadoop job...
15/05/08 02:35:59 INFO impl.YarnClientImpl: Killed application application_1431052132967_0001
Killed.
[root@sandbox h2o-dev-0.3.0.1188-hdp2.2]#
The H2O launch failed because more memory was requested than was available. Make sure you are not trying to specify more memory in the launch parameters than you have available.
How does the architecture of H2O work?
This PDF includes diagrams and slides depicting how H2O works in big data environments.
I received the following error message when launching H2O - how do I resolve the error?
Invalid flow_dir illegal character at index 12...
This error message means that there is a space (or other unsupported character) in your H2O directory. To resolve this error:
- Create a new folder without unsupported characters to use as the H2O
directory (for example,
C:\h2o).
or
- Specify a different save directory using the
-flow_dirparameter when launching H2O:java -jar h2o.jar -flow_dir test
How does ``importFiles()`` work in H2O?
importFiles() gets the basic information for the file and then
returns a key representing that file. This key is used during parsing to
read in the file and to save space so that the file isn’t loaded every
time; instead, it is loaded into H2O then referenced using the key. For
files hosted online, H2O verifies the destination is valid, creates a
vec that loads the file when necessary, and returns a key.
Does H2O support GPUs?
Currently, we do not support this capability. If you are interested in contributing your efforts to support this feature to our open-source code database, please contact us at h2ostream@googlegroups.com.
How can I continue working on a model in H2O after restarting?
There are a number of ways you can save your model in H2O:
- In the web UI, click the Flow menu then click Save Flow. Your flow is saved to the Flows tab in the Help sidebar on the right.
- In the web UI, click the Flow menu then click Download this Flow.... Depending on your browser and configuration, your flow is saved to the “Downloads” folder (by default) or to the location you specify in the pop-up Save As window if it appears.
- (For DRF, GBM, and DL models only): Use model checkpointing to resume
training a model. Copy the
model_idnumber from a built model and paste it into the checkpoint field in thebuildModelcell.
How can I find out more about H2O’s real-time, nano-fast scoring engine?
H2O’s scoring engine uses a Plain Old Java Object (POJO). The POJO code runs quickly but is single-threaded. It is intended for embedding into lightweight real-time environments.
All the work is done by the call to the appropriate predict method. There is no involvement from H2O in this case.
To compare multiple models simultaneously, use the POJO to call the models using multiple threads. For more information on using POJOs, refer to the POJO Quick Start Guide and POJO Java Documentation
In-H2O scoring is triggered on an existing H2O cluster, typically using
a REST API call. H2O evaluates the predictions in a parallel and
distributed fashion for this case. The predictions are stored into a new
Frame and can be written out using h2o.exportFile(), for example.
I am writing an academic research paper and I would like to cite H2O in my bibliography - how should I do that?
To cite our software:
- The H2O.ai Team. (2015) h2o: R Interface for H2O. R package version 3.1.0.99999. http://www.h2o.ai.
- The H2O.ai Team. (2015) h2o: h2o: Python Interface for H2O. Python package version 3.1.0.99999. http://www.h2o.ai.
- The H2O.ai Team. (2015) H2O: Scalable Machine Learning. Version 3.1.0.99999. http://www.h2o.ai.
To cite one of our booklets:
- Nykodym, T., Hussami, N., Kraljevic, T.,Rao, A., and Wang, A. (Sept. 2015). Generalized Linear Modeling with H2O. http://h2o.ai/resources.
- Candel, A., LeDell, E., Parmar, V., and Arora, A. (Sept. 2015). Deep Learning with H2O. http://h2o.ai/resources.
- Click, C., Malohlava, M., Parmar, V., and Roark, H. (Sept. 2015). Gradient Boosted Models with H2O. http://h2o.ai/resources.
- Aiello, S., Eckstrand, E., Fu, A., Landry, M., and Aboyoun, P. (Sept. 2015) Fast Scalable R with H2O. http://h2o.ai/resources.
- Aiello, S., Click, C., Roark, H. and Rehak, L. (Sept. 2015) Machine Learning with Python and H2O http://h2o.ai/resources.
- Malohlava, M., and Tellez, A. (Sept. 2015) Machine Learning with Sparkling Water: H2O + Spark http://h2o.ai/resources.
If you are using Bibtex:
@Manual{h2o_GLM_booklet,
title = {Generalized Linear Modeling with H2O},
author = {Nykodym, T. and Hussami, N. and Kraljevic, T. and Rao, A. and Wang, A.},
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_DL_booklet,
title = {Deep Learning with H2O},
author = {Candel, A. and LeDell, E. and Arora, A. and Parmar, V.},
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_GBM_booklet,
title = {Gradient Boosted Models},
author = {Click, C. and Lanford, J. and Malohlava, M. and Parmar, V. and Roark, H.},
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_R_booklet,
title = {Fast Scalable R with H2O},
author = {Aiello, S. and Eckstrand, E. and Fu, A. and Landry, M. and Aboyoun, P. },
year = {2015},
month = {September},
url = {http://h2o.ai/resources},
}
@Manual{h2o_R_package,
title = {h2o: R Interface for H2O},
author = {The H2O.ai team},
year = {2015},
note = {R package version 3.1.0.99999},
url = {http://www.h2o.ai},
}
@Manual{h2o_Python_module,
title = {h2o: Python Interface for H2O},
author = {The H2O.ai team},
year = {2015},
note = {Python package version 3.1.0.99999},
url = {http://www.h2o.ai},
}
@Manual{h2o_Java_software,
title = {H2O: Scalable Machine Learning},
author = {The H2O.ai team},
year = {2015},
note = {version 3.1.0.99999},
url = {http://www.h2o.ai},
}
How can I use Flow to export the prediction results with a dataset?
After obtaining your results, click the Combine predictions with frame button, then click the View Frame button.
What are these RTMP and py_ temporary Frames? Why are they the same size as my original data?
No data is copied. H2O does a classic copy-on-write optimization. That Frame you see - it’s nothing more than a thin wrapper over an internal list of columns; the columns are shared to avoid the copying.
The RTMP’s now need to be entirely managed by the H2O wrapper - because indeed they are using shared state under the hood. If you delete one, you probably delete parts of others. Instead, temp management should be automatic and “good” - as in: it’s a bug if you need to delete a temp manually, or if passing around Frames, or adding or removing columns turns into large data copies.
R’s GC is now used to remove unused R temps, and when the last use of a shared column goes away, then the H2O wrapper will tell the H2O cluster to remove that no longer needed column.
In other words: Don’t delete RTMPs, they’ll disappear at the next R GC. Don’t worry about copies (they aren’t getting made). Do Nothing and All Is Well.
Hadoop¶
Why did I get an error in R when I tried to save my model to my home directory in Hadoop?
To save the model in HDFS, prepend the save directory with hdfs://:
# build model
model = h2o.glm(model params)
# save model
hdfs_name_node <- "mr-0x6"
hdfs_tmp_dir <- "/tmp/runit”
model_path <- sprintf("hdfs://%s%s", hdfs_name_node, hdfs_tmp_dir)
h2o.saveModel(model, dir = model_path, name = “mymodel")
How do I specify which nodes should run H2O in a Hadoop cluster?
After creating and applying the desired node labels and associating them with specific queues as described in the Hadoop documentation, launch H2O using the following command:
hadoop jar h2odriver.jar -Dmapreduce.job.queuename=<my-h2o-queue> -nodes <num-nodes> -mapperXmx 6g -output hdfsOutputDirName
-Dmapreduce.job.queuename=<my-h2o-queue>represents the queue name-nodes <num-nodes>represents the number of nodes-mapperXmx 6glaunches H2O with 6g of memory-output hdfsOutputDirNamespecifies the HDFS output directory ashdfsOutputDirName
How does H2O handle UDP packet failures? Does H2O quit or retry?
In standard settings, H2O only uses UDP for cloud forming and only if you do not provide a flat file. All other communication is done via TCP. Cloud forming with no flat file is done by repeated broadcasts that are repeated until the cloud forms.
How do I import data from HDFS in R and in Flow?
To import from HDFS in R:
h2o.importFolder(path, pattern = "", destination_frame = "", parse = TRUE, header = NA, sep = "", col.names = NULL, na.strings = NULL)
Here is another example:
# pathToAirlines <- "hdfs://mr-0xd6.0xdata.loc/datasets/airlines_all.csv"
# airlines.hex <- h2o.importFile(path = pathToAirlines, destination_frame = "airlines.hex")
In Flow, the easiest way is to let the auto-suggestion feature in the Search: field complete the path for you. Just start typing the path to the file, starting with the top-level directory, and H2O provides a list of matching files.
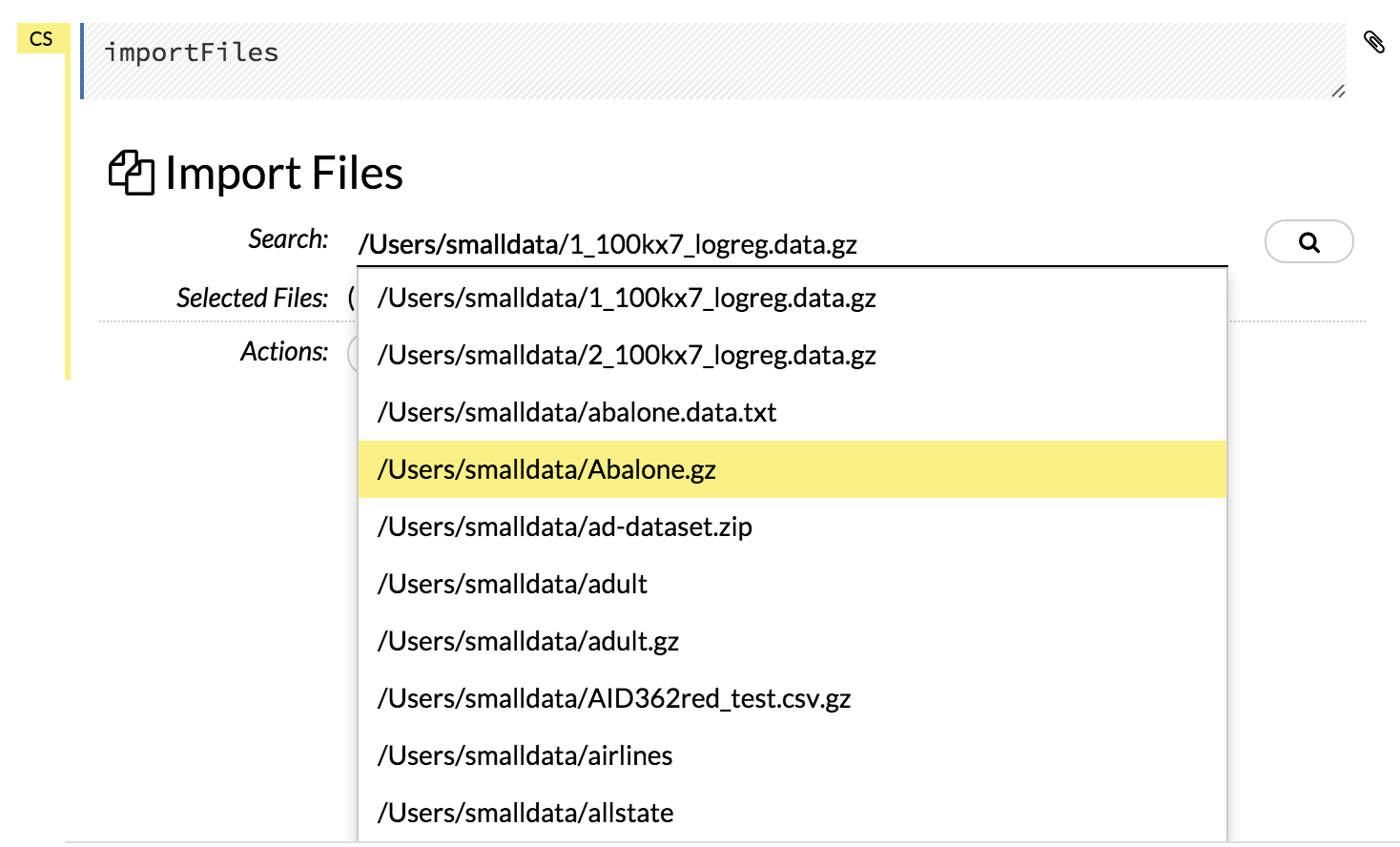
Click the file to add it to the Search: field.
Why do I receive the following error when I try to save my notebook in Flow?
Error saving notebook: Error calling POST /3/NodePersistentStorage/notebook/Test%201 with opts
When you are running H2O on Hadoop, H2O tries to determine the home HDFS
directory so it can use that as the download location. If the default
home HDFS directory is not found, manually set the download location
from the command line using the -flow_dir parameter (for example,
hadoop jar h2odriver.jar <...> -flow_dir hdfs:///user/yourname/yourflowdir).
You can view the default download directory in the logs by clicking
Admin > View logs... and looking for the line that begins
Flow dir:.
How do I access data in HDFS without launching H2O on YARN?
Each h2odriver.jar file is built with a specific Hadoop distribution so in order to have a working HDFS connection download the h2odriver.jar file for your Hadoop distribution from here.
Then run the command to launch the H2O Application in the driver by specifying the classpath:
unzip h2o-<version>.zip
cd h2o-<version>
java -cp h2odriver.jar water.H2OApp
Java¶
How do I use H2O with Java?
There are two ways to use H2O with Java. The simplest way is to call the REST API from your Java program to a remote cluster and should meet the needs of most users.
You can access the REST API documentation within Flow, or on our documentation site.
Flow, Python, and R all rely on the REST API to run H2O. For example, each action in Flow translates into one or more REST API calls. The script fragments in the cells in Flow are essentially the payloads for the REST API calls. Most R and Python API calls translate into a single REST API call.
To see how the REST API is used with H2O:
- Using Chrome as your internet browser, open the developer tab while viewing the web UI. As you perform tasks, review the network calls made by Flow.
- Write an R program for H2O using the H2O R package that uses
h2o.startLogging()at the beginning. All REST API calls used are logged.
The second way to use H2O with Java is to embed H2O within your Java application, similar to Sparkling Water.
How do I communicate with a remote cluster using the REST API?
To create a set of bare POJOs for the REST API payloads that can be used by JVM REST API clients:
- Clone the sources from GitHub.
- Start an H2O instance.
- Enter
% cd py. - Enter
% python generate_java_binding.py.
This script connects to the server, gets all the metadata for the REST
API schemas, and writes the Java POJOs to
{sourcehome}/build/bindings/Java.
I keep getting a message that I need to install Java. I have the latest version of Java installed, but I am still getting this message. What should I do?
This error message displays if the JAVA_HOME environment variable is
not set correctly. The JAVA_HOME variable is likely points to Apple
Java version 6 instead of Oracle Java version 8.
If you are running OS X 10.7 or earlier, enter the following in
Terminal:
export JAVA_HOME=/Library/Internet\ Plug-Ins/JavaAppletPlugin.plugin/Contents/Home
If you are running OS X 10.8 or later, modify the launchd.plist by entering the following in Terminal:
cat << EOF | sudo tee /Library/LaunchDaemons/setenv.JAVA_HOME.plist
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE plist PUBLIC "-//Apple//DTD PLIST 1.0//EN" "http://www.apple.com/DTDs/PropertyList-1.0.dtd">
<plist version="1.0">
<dict>
<key>Label</key>
<string>setenv.JAVA_HOME</string>
<key>ProgramArguments</key>
<array>
<string>/bin/launchctl</string>
<string>setenv</string>
<string>JAVA_HOME</string>
<string>/Library/Internet Plug-Ins/JavaAppletPlugin.plugin/Contents/Home</string>
</array>
<key>RunAtLoad</key>
<true/>
<key>ServiceIPC</key>
<false/>
</dict>
</plist>
EOF
Python¶
I tried to install H2O in Python but ``pip install scikit-learn`` failed - what should I do?
Use the following commands (prepending with sudo if necessary):
easy_install pip
pip install numpy
brew install gcc
pip install scipy
pip install scikit-learn
If you are still encountering errors and you are using OSX, the default version of Python may be installed. We recommend installing the Homebrew version of Python instead:
brew install python
If you are encountering errors related to missing Python packages when using H2O, refer to the following list for a complete list of all Python packages, including dependencies:
griptabulatewheelejsonliteipythonnumpyscipypandas-U gensimjupyter-U PILnltkbeautifulsoup4
How do I specify a value as an enum in Python? Is there a Python equivalent of ``as.factor()`` in R?
Use .asfactor() to specify a value as an enum.
I received the following error when I tried to install H2O using the Python instructions on the downloads page - what should I do to resolve it?
Downloading/unpacking http://h2o-release.s3.amazonaws.com/h2o/rel-shannon/12/Python/h2o-3.0.0.12-py2.py3-none-any.whl
Downloading h2o-3.0.0.12-py2.py3-none-any.whl (43.1Mb): 43.1Mb downloaded
Running setup.py egg_info for package from http://h2o-release.s3.amazonaws.com/h2o/rel-shannon/12/Python/h2o-3.0.0.12-py2.py3-none-any.whl
Traceback (most recent call last):
File "<string>", line 14, in <module>
IOError: [Errno 2] No such file or directory: '/tmp/pip-nTu3HK-build/setup.py'
Complete output from command python setup.py egg_info:
Traceback (most recent call last):
File "<string>", line 14, in <module>
IOError: [Errno 2] No such file or directory: '/tmp/pip-nTu3HK-build/setup.py'
---
Command python setup.py egg_info failed with error code 1 in /tmp/pip-nTu3HK-build
With Python, there is no automatic update of installed packages, so you
must upgrade manually. Additionally, the package distribution method
recently changed from distutils to wheel. The following
procedure should be tried first if you are having trouble installing the
H2O package, particularly if error messages related to bdist_wheel
or eggs display.
# this gets the latest setuptools
# see https://pip.pypa.io/en/latest/installing.html
wget https://bootstrap.pypa.io/ez_setup.py -O - | sudo python
# platform dependent ways of installing pip are at
# https://pip.pypa.io/en/latest/installing.html
# but the above should work on most linux platforms?
# on ubuntu
# if you already have some version of pip, you can skip this.
sudo apt-get install python-pip
# the package manager doesn't install the latest. upgrade to latest
# we're not using easy_install any more, so don't care about checking that
pip install pip --upgrade
# I've seen pip not install to the final version ..i.e. it goes to an almost
# final version first, then another upgrade gets it to the final version.
# We'll cover that, and also double check the install.
# after upgrading pip, the path name may change from /usr/bin to /usr/local/bin
# start a new shell, just to make sure you see any path changes
bash
# Also: I like double checking that the install is bulletproof by reinstalling.
# Sometimes it seems like things say they are installed, but have errors during the install. Check for no errors or stack traces.
pip install pip --upgrade --force-reinstall
# distribute should be at the most recent now. Just in case
# don't do --force-reinstall here, it causes an issue.
pip install distribute --upgrade
# Now check the versions
pip list | egrep '(distribute|pip|setuptools)'
distribute (0.7.3)
pip (7.0.3)
setuptools (17.0)
# Re-install wheel
pip install wheel --upgrade --force-reinstall
After completing this procedure, go to Python and use h2o.init() to
start H2O in Python.
Notes:
If you use gradlew to build the jar yourself, you have to start the jar >yourself before you do
h2o.init().If you download the jar and the H2O package,
h2o.init()will work like R >and you don’t have to start the jar yourself.
How should I specify the datatype during import in Python?
Refer to the following example:
#Let's say you want to change the second column "CAPSULE" of prostate.csv
#to categorical. You have 3 options.
#Option 1. Use a dictionary of column names to types.
fr = h2o.import_file("smalldata/logreg/prostate.csv", col_types = {"CAPSULE":"Enum"})
fr.describe()
#Option 2. Use a list of column types.
c_types = [None]*9
c_types[1] = "Enum"
fr = h2o.import_file("smalldata/logreg/prostate.csv", col_types = c_types)
fr.describe()
#Option 3. Use parse_setup().
fraw = h2o.import_file("smalldata/logreg/prostate.csv", parse = False)
fsetup = h2o.parse_setup(fraw)
fsetup["column_types"][1] = '"Enum"'
fr = h2o.parse_raw(fsetup)
fr.describe()
How do I view a list of variable importances in Python?
Use model.varimp(return_list=True) as shown in the following
example:
model = h2o.gbm(y = "IsDepDelayed", x = ["Month"], training_frame = df)
vi = model.varimp(return_list=True)
Out[26]:
[(u'Month', 69.27436828613281, 1.0, 1.0)]
What is PySparkling? How can I use it for grid search or early stopping?
PySparkling basically calls H2O Python functions for all operations on H2O data frames. You can perform all H2O Python operations available in H2O Python version 3.6.0.3 or later from PySparkling.
For help on a function within IPython Notebook, run H2OGridSearch?
Here is an example of grid search in PySparkling:
from h2o.grid.grid_search import H2OGridSearch
from h2o.estimators.gbm import H2OGradientBoostingEstimator
iris = h2o.import_file("/Users/nidhimehta/h2o-dev/smalldata/iris/iris.csv")
ntrees_opt = [5, 10, 15]
max_depth_opt = [2, 3, 4]
learn_rate_opt = [0.1, 0.2]
hyper_parameters = {"ntrees": ntrees_opt, "max_depth":max_depth_opt,
"learn_rate":learn_rate_opt}
gs = H2OGridSearch(H2OGradientBoostingEstimator(distribution='multinomial'), hyper_parameters)
gs.train(x=range(0,iris.ncol-1), y=iris.ncol-1, training_frame=iris, nfold=10)
#gs.show
print gs.sort_by('logloss', increasing=True)
Here is an example of early stopping in PySparkling:
from h2o.grid.grid_search import H2OGridSearch
from h2o.estimators.deeplearning import H2ODeepLearningEstimator
hidden_opt = [[32,32],[32,16,8],[100]]
l1_opt = [1e-4,1e-3]
hyper_parameters = {"hidden":hidden_opt, "l1":l1_opt}
model_grid = H2OGridSearch(H2ODeepLearningEstimator, hyper_params=hyper_parameters)
model_grid.train(x=x, y=y, distribution="multinomial", epochs=1000, training_frame=train,
validation_frame=test, score_interval=2, stopping_rounds=3, stopping_tolerance=0.05, stopping_metric="misclassification")
Do you have a tutorial for grid search in Python?
Yes, a notebook is available here that demonstrates the use of grid search in Python.
R¶
Which versions of R are compatible with H2O?
Currently, the only version of R that is known to not work well with H2O is R version 3.1.0 (codename “Spring Dance”). If you are using this version, we recommend upgrading the R version before using H2O.
What R packages are required to use H2O?
The following packages are required:
methodsstatmodstatsgraphicsRCurljsonlitetoolsutils
Some of these packages have dependencies; for example, bitops is
required, but it is a dependency of the RCurl package, so bitops
is automatically included when RCurl is installed.
If you are encountering errors related to missing R packages when using H2O, refer to the following list for a complete list of all R packages, including dependencies:
statmodbitopsRCurljsonlitemethodsstatsgraphicstoolsutilsstringimagrittrcolorspacestringrRColorBrewerdichromatmunselllabelingplyrdigestgtablereshape2scalesprotoggplot2h2oEnsemblegtoolsgdatacaToolsgplotschronROCRdata.tablecvAUC
Finally, if you are running R on Linux, then you must install libcurl, which allows H2O to communicate with R.
How can I install the H2O R package if I am having permissions problems?
This issue typically occurs for Linux users when the R software was installed by a root user. For more information, refer to the following link.
To specify the installation location for the R packages, create a file
that contains the R_LIBS_USER environment variable:
echo R_LIBS_USER=\"~/.Rlibrary\" > ~/.Renviron
Confirm the file was created successfully using cat:
$ cat ~/.Renviron
You should see the following output:
R_LIBS_USER="~/.Rlibrary"
Create a new directory for the environment variable:
$ mkdir ~/.Rlibrary
Start R and enter the following:
.libPaths()
Look for the following output to confirm the changes:
[1] "<Your home directory>/.Rlibrary"
[2] "/Library/Frameworks/R.framework/Versions/3.1/Resources/library"
I received the following error message after launching H2O in RStudio and using ``h2o.init`` - what should I do to resolve this error?
Error in h2o.init() :
Version mismatch! H2O is running version 3.2.0.9 but R package is version 3.2.0.3
This error is due to a version mismatch between the H2O R package and the running H2O instance. Make sure you are using the latest version of both files by downloading H2O from the downloads page and installing the latest version and that you have removed any previous H2O R package versions by running:
if ("package:h2o" %in% search()) { detach("package:h2o", unload=TRUE) }
if ("h2o" %in% rownames(installed.packages())) { remove.packages("h2o") }
Make sure to install the dependencies for the H2O R package as well:
if (! ("methods" %in% rownames(installed.packages()))) { install.packages("methods") }
if (! ("statmod" %in% rownames(installed.packages()))) { install.packages("statmod") }
if (! ("stats" %in% rownames(installed.packages()))) { install.packages("stats") }
if (! ("graphics" %in% rownames(installed.packages()))) { install.packages("graphics") }
if (! ("RCurl" %in% rownames(installed.packages()))) { install.packages("RCurl") }
if (! ("jsonlite" %in% rownames(installed.packages()))) { install.packages("jsonlite") }
if (! ("tools" %in% rownames(installed.packages()))) { install.packages("tools") }
if (! ("utils" %in% rownames(installed.packages()))) { install.packages("utils") }
Finally, install the latest version of the H2O package for R:
install.packages("h2o", type="source", repos=(c("http://h2o-release.s3.amazonaws.com/h2o/<branch_name>/<build_number>/R")))
library(h2o)
localH2O = h2o.init(nthreads=-1)
If your R version is older than the H2O R package, upgrade your R
version using update.packages(checkBuilt=TRUE, ask=FALSE).
I received the following error message after trying to run some code - what should I do?
> fit <- h2o.deeplearning(x=2:4, y=1, training_frame=train_hex)
|=========================================================================================================| 100%
Error in model$training_metrics$MSE :
$ operator not defined for this S4 class
In addition: Warning message:
Not all shim outputs are fully supported, please see ?h2o.shim for more information
Remove the h2o.shim(enable=TRUE) line and try running the code
again. Note that the h2o.shim is only a way to notify users of
previous versions of H2O about changes to the H2O R package - it will
not revise your code, but provides suggested replacements for deprecated
commands and parameters.
How do I extract the model weights from a model I’ve creating using H2O in R? I’ve enabled ``extract_model_weights_and_biases``, but the output refers to a file I can’t open in R.
For an example of how to extract weights and biases from a model, refer to the following repo location on GitHub.
How do I extract the run time of my model as output?
For the following example:
out.h2o.rf = h2o.randomForest( x=c("x1", "x2", "x3", "w"), y="y", training_frame=h2o.df.train, seed=555, model_id= "my.model.1st.try.out.h2o.rf" )
Use out.h2o.rf@model$run_time to determine the value of the
run_time variable.
What is the best way to do group summarizations? For example, getting sums of specific columns grouped by a categorical column.
We strongly recommend using h2o.group_by for this function instead
of h2o.ddply, as shown in the following example:
newframe <- h2o.group_by(h2oframe, by="footwear_category", nrow("email_event_click_ct"), sum("email_event_click_ct"), mean("email_event_click_ct"), sd("email_event_click_ct"), gb.control = list( col.names=c("count", "total_email_event_click_ct", "avg_email_event_click_ct", "std_email_event_click_ct") ) )
Using gb.control is optional; here it is included so the column
names are user-configurable.
The by option can take a list of columns if you want to group by
more than one column to compute the summary as shown in the following
example:
newframe <- h2o.group_by(h2oframe, by=c("footwear_category","age_group"), nrow("email_event_click_ct"), sum("email_event_click_ct"), mean("email_event_click_ct"), sd("email_event_click_ct"), gb.control = list( col.names=c("count", "total_email_event_click_ct", "avg_email_event_click_ct", "std_email_event_click_ct") ) )
I’m using Linux and I want to run H2O in R - are there any dependencies I need to install?
Yes, make sure to install libcurl, which allows H2O to communicate
with R. We also recommend disabling SElinux and any firewalls, at least
initially until you have confirmed H2O can initialize.
- On Ubuntu, run:
apt-get install libcurl4-openssl-dev - On CentOS, run:
yum install libcurl-devel
How do I change variable/header names on an H2O frame in R?
There are two ways to change header names. To specify the headers during parsing, import the headers in R and then specify the header as the column name when the actual data frame is imported:
header <- h2o.importFile(path = pathToHeader)
data <- h2o.importFile(path = pathToData, col.names = header)
data
You can also use the names() function:
header <- c("user", "specified", "column", "names")
data <- h2o.importFile(path = pathToData)
names(data) <- header
To replace specific column names, you can also use a sub/gsub in R:
header <- c("user", "specified", "column", "names")
## I want to replace "user" column with "computer"
data <- h2o.importFile(path = pathToData)
names(data) <- sub(pattern = "user", replacement = "computer", x = names(header))
My R terminal crashed - how can I re-access my H2O frame?
Launch H2O and use your web browser to access the web UI, Flow, at
localhost:54321. Click the Data menu, then click List All
Frames. Copy the frame ID, then run h2o.ls() in R to list all the
frames, or use the frame ID in the following code (replacing
YOUR_FRAME_ID with the frame ID):
library(h2o)
localH2O = h2o.init(ip="sri.h2o.ai", port=54321, startH2O = F, strict_version_check=T)
data_frame <- h2o.getFrame(frame_id = "YOUR_FRAME_ID")
How do I remove rows containing NAs in an H2OFrame?
To remove NAs from rows:
a b c d e
1 0 NA NA NA NA
2 0 2 2 2 2
3 0 NA NA NA NA
4 0 NA NA 1 2
5 0 NA NA NA NA
6 0 1 2 3 2
Removing rows 1, 3, 4, 5 to get:
a b c d e
2 0 2 2 2 2
6 0 1 2 3 2
Use na.omit(myFrame), where myFrame represents the name of the
frame you are editing.
I installed H2O in R using OS X and updated all the dependencies, but the following error message displayed: ``Error in .h2o.doSafeREST(h2oRestApiVersion = h2oRestApiVersion, Unexpected CURL error: Empty reply from server`` - what should I do?
This error message displays if the JAVA_HOME environment variable is
not set correctly. The JAVA_HOME variable is likely points to Apple
Java version 6 instead of Oracle Java version 8.
If you are running OS X 10.7 or earlier, enter the following in
Terminal:
export JAVA_HOME=/Library/Internet\ Plug-Ins/JavaAppletPlugin.plugin/Contents/Home
If you are running OS X 10.8 or later, modify the launchd.plist by entering the following in Terminal:
cat << EOF | sudo tee /Library/LaunchDaemons/setenv.JAVA_HOME.plist
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE plist PUBLIC "-//Apple//DTD PLIST 1.0//EN" "http://www.apple.com/DTDs/PropertyList-1.0.dtd">
<plist version="1.0">
<dict>
<key>Label</key>
<string>setenv.JAVA_HOME</string>
<key>ProgramArguments</key>
<array>
<string>/bin/launchctl</string>
<string>setenv</string>
<string>JAVA_HOME</string>
<string>/Library/Internet Plug-Ins/JavaAppletPlugin.plugin/Contents/Home</string>
</array>
<key>RunAtLoad</key>
<true/>
<key>ServiceIPC</key>
<false/>
</dict>
</plist>
EOF
How does the ``col.names`` argument work in ``group_by``?
You need to add the col.names inside the gb.control list. Refer
to the following example:
newframe <- h2o.group_by(dd, by="footwear_category", nrow("email_event_click_ct"), sum("email_event_click_ct"), mean("email_event_click_ct"),
sd("email_event_click_ct"), gb.control = list( col.names=c("count", "total_email_event_click_ct", "avg_email_event_click_ct", "std_email_event_click_ct") ) )
newframe$avg_email_event_click_ct2 = newframe$total_email_event_click_ct / newframe$count
How are the results of ``h2o.predict`` displayed?
The order of the rows in the results for h2o.predict is the same as
the order in which the data was loaded, even if some rows fail (for
example, due to missing values or unseen factor levels). To bind a
per-row identifier, use cbind.
How do I view all the variable importances for a model?
By default, H2O returns the top five and lowest five variable importances. To view all the variable importances, use the following:
model <- h2o.getModel(model_id = "my_H2O_modelID",conn=localH2O)
varimp<-as.data.frame(h2o.varimp(model))
How do I add random noise to a column in an H2O frame?
To add random noise to a column in an H2O frame, refer to the following example:
h2o.init()
fr <- as.h2o(iris)
|======================================================================| 100%
random_column <- h2o.runif(fr)
new_fr <- h2o.cbind(fr,random_column)
new_fr
Sparkling Water¶
What is Sparkling Water?
Sparkling Water allows users to combine the fast, scalable machine learning algorithms of H2O with the capabilities of Spark. With Sparkling Water, users can drive computation from Scala/R/Python and utilize the H2O Flow UI, providing an ideal machine learning platform for application developers.
What are the advantages of using Sparkling Water compared with H2O?
Sparkling Water contains the same features and functionality as H2O but provides a way to use H2O with Spark, a large-scale cluster framework.
Sparkling Water is ideal for H2O users who need to manage large clusters for their data processing needs and want to transfer data from Spark to H2O (or vice versa).
There is also a Python interface available to enable access to Sparkling Water directly from PySpark.
How do I filter an H2OFrame using Sparkling Water?
Filtering columns is easy: just remove the unnecessary columns or create
a new H2OFrame from the columns you want to include
(Frame(String[] names, Vec[] vec)), then make the H2OFrame wrapper
around it (new H2OFrame(frame)).
Filtering rows is a little bit harder. There are two ways:
- Create an additional binary vector holding
1/0for thein/outsample (make sure to take this additional vector into account in your computations). This solution is quite cheap, since you do not duplicate data - just create a simple vector in a data walk.
or
- Create a new frame with the filtered rows. This is a harder task,
since you have to copy data. For reference, look at the #deepSlice
call on Frame (
H2OFrame)
How can I save and load a K-means model using Sparkling Water?
The following example code defines the save and load functions explicitly.
import water._
import _root_.hex._
import java.net.URI
import water.serial.ObjectTreeBinarySerializer
// Save H2O model (as binary)
def exportH2OModel(model : Model[_,_,_], destination: URI): URI = {
val modelKey = model._key.asInstanceOf[Key[_ <: Keyed[_ <: Keyed[_ <: AnyRef]]]]
val keysToExport = model.getPublishedKeys()
// Prepend model key
keysToExport.add(0, modelKey)
new ObjectTreeBinarySerializer().save(keysToExport, destination)
destination
}
// Get model from H2O DKV and Save to disk
val gbmModel: _root_.hex.tree.gbm.GBMModel = DKV.getGet("model")
exportH2OModel(gbmModel, new File("../h2omodel.bin").toURI)
def loadH2OModel[M <: Model[_, _, _]](source: URI) : M = {
val l = new ObjectTreeBinarySerializer().load(source)
l.get(0).get().asInstanceOf[M]
}
// Load H2O model
def loadH2OModel[M <: Model[_, _, _]](source: URI) : M = {
val l = new ObjectTreeBinarySerializer().load(source)
l.get(0).get().asInstanceOf[M]
}
// Load model
val h2oModel: Model[_, _, _] = loadH2OModel(new File("../h2omodel.bin").toURI)
How do I inspect H2O using Flow while a droplet is running?
If your droplet execution time is very short, add a simple sleep statement to your code:
Thread.sleep(...)
How do I change the memory size of the executors in a droplet?
There are two ways to do this:
- Change your default Spark setup in
$SPARK_HOME/conf/spark-defaults.conf
or
- Pass
--confvia spark-submit when you launch your droplet (e.g.,
$SPARK_HOME/bin/spark-submit --conf spark.executor.memory=4g --master $MASTER --class org.my.Droplet $TOPDIR/assembly/build/libs/droplet.jar
I received the following error while running Sparkling Water using multiple nodes, but not when using a single node - what should I do?
onExCompletion for water.parser.ParseDataset$MultiFileParseTask@31cd4150
water.DException$DistributedException: from /10.23.36.177:54321; by class water.parser.ParseDataset$MultiFileParseTask; class water.DException$DistributedException: from /10.23.36.177:54325; by class water.parser.ParseDataset$MultiFileParseTask; class water.DException$DistributedException: from /10.23.36.178:54325; by class water.parser.ParseDataset$MultiFileParseTask$DistributedParse; class java.lang.NullPointerException: null
at water.persist.PersistManager.load(PersistManager.java:141)
at water.Value.loadPersist(Value.java:226)
at water.Value.memOrLoad(Value.java:123)
at water.Value.get(Value.java:137)
at water.fvec.Vec.chunkForChunkIdx(Vec.java:794)
at water.fvec.ByteVec.chunkForChunkIdx(ByteVec.java:18)
at water.fvec.ByteVec.chunkForChunkIdx(ByteVec.java:14)
at water.MRTask.compute2(MRTask.java:426)
at water.MRTask.compute2(MRTask.java:398)
This error output displays if the input file is not present on all nodes. Because of the way that Sparkling Water distributes data, the input file is required on all nodes (including remote), not just the primary node. Make sure there is a copy of the input file on all the nodes, then try again.
Are there any drawbacks to using Sparkling Water compared to standalone H2O?
The version of H2O embedded in Sparkling Water is the same as the standalone version.
How do I use Sparkling Water from the Spark shell?
There are two methods:
- Use
$SPARK_HOME/bin/spark-shell --packages ai.h2o:sparkling-water-core_2.10:1.3.3
or
bin/sparkling-shell
The software distribution provides example scripts in the
examples/scripts directory:
bin/sparkling-shell -i examples/scripts/chicagoCrimeSmallShell.script.scala
For either method, initialize H2O as shown in the following example:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc).start()
After successfully launching H2O, the following output displays:
Sparkling Water Context:
* number of executors: 3
* list of used executors:
(executorId, host, port)
------------------------
(1,Michals-MBP.0xdata.loc,54325)
(0,Michals-MBP.0xdata.loc,54321)
(2,Michals-MBP.0xdata.loc,54323)
------------------------
Open H2O Flow in browser: http://172.16.2.223:54327 (CMD + click in Mac OSX)
How do I use H2O with Spark Submit?
Spark Submit is for submitting self-contained applications. For more information, refer to the Spark documentation.
First, initialize H2O:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc).start()
The Sparkling Water distribution provides several examples of self-contained applications built with Sparkling Water. To run the examples:
bin/run-example.sh ChicagoCrimeAppSmall
The “magic” behind run-example.sh is a regular Spark Submit:
$SPARK_HOME/bin/spark-submit ChicagoCrimeAppSmall --packages ai.h2o:sparkling-water-core_2.10:1.3.3 --packages ai.h2o:sparkling-water-examples_2.10:1.3.3
How do I use Sparkling Water with Databricks cloud?
Sparkling Water compatibility with Databricks cloud is still in development.
How do I develop applications with Sparkling Water?
For a regular Spark application (a self-contained application as
described in the Spark
documentation),
the app needs to initialize H2OServices via H2OContext:
import org.apache.spark.h2o._
val h2oContext = new H2OContext(sc).start()
For more information, refer to the Sparkling Water development documentation.
How do I connect to Sparkling Water from R or Python?
After starting H2OServices by starting H2OContext, point your
client to the IP address and port number specified in H2OContext.
Tunneling between servers with H2O¶
To tunnel between servers (for example, due to firewalls):
- Use ssh to log in to the machine where H2O will run.
- Start an instance of H2O by locating the working directory and calling a java command similar to the following example.
The port number chosen here is arbitrary; yours may be different.
$ java -jar h2o.jar -port 55599
This returns output similar to the following:
irene@mr-0x3:~/target$ java -jar h2o.jar -port 55599
04:48:58.053 main INFO WATER: ----- H2O started -----
04:48:58.055 main INFO WATER: Build git branch: master
04:48:58.055 main INFO WATER: Build git hash: 64fe68c59ced5875ac6bac26a784ce210ef9f7a0
04:48:58.055 main INFO WATER: Build git describe: 64fe68c
04:48:58.055 main INFO WATER: Build project version: 1.7.0.99999
04:48:58.055 main INFO WATER: Built by: 'Irene'
04:48:58.055 main INFO WATER: Built on: 'Wed Sep 4 07:30:45 PDT 2013'
04:48:58.055 main INFO WATER: Java availableProcessors: 4
04:48:58.059 main INFO WATER: Java heap totalMemory: 0.47 gb
04:48:58.059 main INFO WATER: Java heap maxMemory: 6.96 gb
04:48:58.060 main INFO WATER: ICE root: '/tmp'
04:48:58.081 main INFO WATER: Internal communication uses port: 55600
+ Listening for HTTP and REST traffic on
+ http://192.168.1.173:55599/
04:48:58.109 main INFO WATER: H2O cloud name: 'irene'
04:48:58.109 main INFO WATER: (v1.7.0.99999) 'irene' on
/192.168.1.173:55599, discovery address /230 .252.255.19:59132
04:48:58.111 main INFO WATER: Cloud of size 1 formed [/192.168.1.173:55599]
04:48:58.247 main INFO WATER: Log dir: '/tmp/h2ologs'
Log into the remote machine where the running instance of H2O will be forwarded using a command similar to the following. (Your specified port numbers and IP address will be different.)
ssh -L 55577:localhost:55599 irene@192.168.1.173Check the cluster status.
You are now using H2O from localhost:55577, but the instance of H2O is running on the remote server (in this case the server with the ip address 192.168.1.xxx) at port number 55599.
To see this in action note that the web UI is pointed at localhost:55577, but that the cluster status shows the cluster running on 192.168.1.173:55599.