R Tutorial¶
The purpose of this tutorial is to give a sample workflow for new users of H2O’s R API. The objective is to learn the basic syntax of H2O, including importing and parsing files, specifying a model, and obtaining model output.
Those who have never used H2O before should see the quick start guide for additional instructions on how to run H2O. The following tutorial will follow the assumption that the user was able to install H2O in R.
Getting Started¶
R uses an REST API to send directives to H2O which means that we need a reference object in R to the H:sub: 2O instance. The user can either start H2O outside of R and connect to it after or the user can launch directly from R and when the R session closes the H2O instance will be killed with it. The client object will later be used to direct R to datasets and models sitting in H2O.
Launch From R
By default if the argument max_mem_size is not specified when running h2o.init() the heap size of the H2O running on 32-bit Java is 1g and on 64-bit Java is 1/4 of the total memory available on the machine. If the user is using a 32-bit version the function will run a check and suggest an upgrade.
library(h2o)
localH2O <- h2o.init(ip = 'localhost', port = 54321, max_mem_size = '4g')
Launch From Command Line
Launch an instance from the command line on your desktop, on ec2 instances, or Hadoop servers by following one of the deployment tutorials. Once the H2O cluster is launched, simply initialize the connection by taking one node in the cluster and run h2o.init with its IP Address and port.
library(h2o)
localH2O <- h2o.init(ip = '192.168.1.161', port =54321)
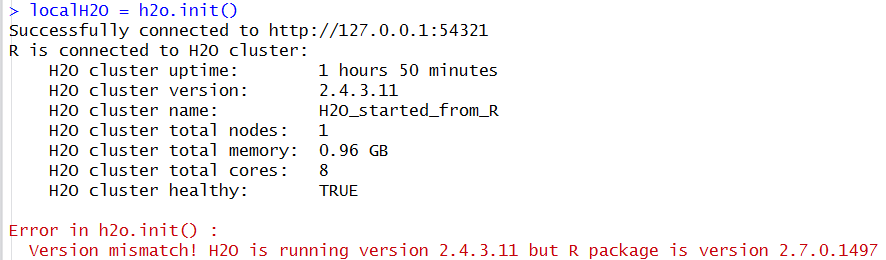
Warning
If the version of H2O instance running is different from the package version loaded in R, a warning message indicating a version mismatch will appear. To fix this issue either update the R package or launch an H2O instance using the jar file from the installed package.
Cluster Info
At any point if the user want to check on the status and health of the H2O cluster h2o.clusterInfo() returns an easy to read list of information on the cluster.
library(h2o)
localH2O = h2o.init(ip = 'localhost', port = 54321)
h2o.clusterInfo(localH2O)
Importing Data¶
Import File
H2O’s package has consolidated all the different import functions supported. Although h2o.importFolder and h2o.importHDFS will still work, it should be noted that these functions are deprecated and should be updated to h2o.importFile.
## To import small iris data file from H\ :sub:`2`\ O's package
irisPath = system.file("extdata", "iris.csv", package="h2o")
iris.hex = h2o.importFile(localH2O, path = irisPath, key = "iris.hex")
summary(iris.hex)
## To import an entire folder of files as one data object
pathToFolder = "/Users/Amy/0xdata/data/airlines/"
airlines.hex = h2o.importFile(localH2O, path = pathToFolder, key = "airlines.hex")
summary(airlines.hex)
## To import from HDFS
pathToData = "hdfs://mr-0xd6.0xdata.loc/datasets/airlines_all.csv"
airlines.hex = h2o.importFile(localH2O, path = pathToData, key = "airlines.hex")
summary(airlines.hex)
Upload File
To upload a file from your local disk you can run upload file, however importFile is recommend if applicable.
irisPath = system.file("extdata", "iris.csv", package="h2o")
iris.hex = h2o.uploadFile(localH2O, path = irisPath, key = "iris.hex")
summary(iris.hex)
Data Manipulation and Description¶
Any Factor
Used to determine if any column in a data set is a factor.
irisPath = system.file("extdata", "iris_wheader.csv", package="h2o")
iris.hex = h2o.importFile(localH2O, path = irisPath)
h2o.anyFactor(iris.hex)
As Data Frame
Used to convert an H2O parsed data object into an R data frame (which can subsequently be manipulated using R calls). While this is frequently useful, as.data.frame should be used with care when converting H2O Parsed Data objects. Data sets that are easily and quickly handled by H2O are often too large to be treated equivalently well in R.
prosPath <- system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath)
prostate.data.frame<- as.data.frame(prostate.hex)
summary(prostate.data.frame)
head(prostate.data.frame)
As Factor
Used to convert an integer into a non-ordered factor (alternatively called an enum or categorical).
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath)
prostate.hex[,4] = as.factor(prostate.hex[,4])
summary(prostate.hex)
As H2O
Used to pass a data frame from inside of the R environment to the H2O instance.
data(iris)
summary(iris)
iris.r <- iris
iris.h2o <- as.h2o(localH2O, iris.r, key="iris.h2o")
class(iris.h2o)
Assign H2O
Used to create an hex key on the server where H2O is running for data sets manipulated in R. For instance, in the example below, the prostate data set was uploaded to the H2O instance, and was manipulated to remove outliers. Saving the new data set on the H2O server so that it can be subsequently be analyzed with H2O without overwriting the original data set relies on h2o.assign.
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath)
prostate.qs = quantile(prostate.hex$PSA)
PSA.outliers = prostate.hex[prostate.hex$PSA <= prostate.qs[2] | prostate.hex$PSA >= prostate.qs[10],]
PSA.outliers = h2o.assign(PSA.outliers, "PSA.outliers")
nrow(prostate.hex)
nrow(PSA.outliers)
Colnames
Used to obtain a list of the column names in a data set.
irisPath = system.file("extdata", "iris.csv", package="h2o")
iris.hex = h2o.importFile(localH2O, path = irisPath, key = "iris.hex")
summary(iris.hex)
colnames(iris.hex)
Extremes
Used to obtain the maximum and minimum values in real valued columns.
ausPath = system.file("extdata", "australia.csv", package="h2o")
australia.hex = h2o.importFile(localH2O, path = ausPath, key = "australia.hex")
min(australia.hex)
min(c(-1, 0.5, 0.2), FALSE, australia.hex[,1:4])
Quantiles
Used to request quantiles for an H2O parsed data set. When requested for a full parsed data set quantiles() returns a matrix displaying quantile information for all numeric columns in the data set.
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath)
quantile(prostate.hex)
Summary
Used to generate an R like summary for each of the columns of a data set. For continuous reals this produces a summary that includes information on quartiles, min, max and mean. For factors this produces information on counts of elements within each factor level. For information on the Summary algorithm see Summary
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath)
summary(prostate.hex)
summary(prostate.hex$GLEASON)
summary(prostate.hex[,4:6])
H2O Table
Used to summarize information in data. Note that because H2O handles such large data sets, it is possible for users to generate tables that are larger that R’s capacity. To minimize this risk and allow users to work uninterrupted, h2o.table is called inside of a call for head() or tail(). Within head() and tail() users can explicitly specify the number of rows in the table to return.
head(h2o.table(prostate.hex[,3]))
head(h2o.table(prostate.hex[,c(3,4)]))
Generate Random Uniformly Distributed Numbers
Runif is used to append a column of random numbers to an H2O data frame and facilitate creating test/ train splits of data for analysis and validation in H2O.
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath, key = "prostate.hex")
s = h2o.runif(prostate.hex)
summary(s)
prostate.train = prostate.hex[s <= 0.8,]
prostate.train = h2o.assign(prostate.train, "prostate.train")
prostate.test = prostate.hex[s > 0.8,]
prostate.test = h2o.assign(prostate.test, "prostate.test")
nrow(prostate.train) + nrow(prostate.test)
Split Frame
The function generates two subset of an existing H2O data set according to user-specified ratios which can be used as test/train sets. This is the preferred method of splitting a data frame as it’s faster and more stable than running runif across entire data however runif can be used for customized frame splitting.
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath, key = "prostate.hex")
prostate.split = h2o.splitFrame(data = prostate.hex , ratios = 0.75)
prostate.train = prostate.split[1]
prostate.test = prostate.split[2]
summary(prostate.train)
summary(prostate.test)
Running Models¶
GBM
Gradient Boosted Models. For information on the GBM algorithm see Gradient Boosted Regression and Classification
ausPath = system.file("extdata", "australia.csv", package="h2o")
australia.hex = h2o.importFile(localH2O, path = ausPath)
independent <- c("premax", "salmax","minairtemp", "maxairtemp",
"maxsst", "maxsoilmoist", "Max_czcs")
dependent <- "runoffnew"
h2o.gbm(y = dependent, x = independent, data = australia.hex,
n.trees = 10, interaction.depth = 3,
n.minobsinnode = 2, shrinkage = 0.2, distribution= "gaussian")
Run multinomial classification GBM on abalone data
h2o.gbm(y = dependent, x = independent, data = australia.hex, n.trees
= 15, interaction.depth = 5,
n.minobsinnode = 2, shrinkage = 0.01, distribution= "multinomial")
GLM
Generalized linear models, which are used to develop linear models for exponential distributions. Regularization can be applied. For information on the GBM algorithm see Generalized Linear Model
prostate.hex = h2o.importFile(localH2O, path =
"https://raw.github.com/0xdata/h2o/master/smalldata/logreg/prostate.csv",
key = "prostate.hex")
h2o.glm(y = "CAPSULE", x = c("AGE","RACE","PSA","DCAPS"), data =
prostate.hex, family = "binomial", nfolds = 10, alpha = 0.5)
myX = setdiff(colnames(prostate.hex), c("ID", "DPROS", "DCAPS", "VOL"))
h2o.glm(y = "VOL", x = myX, data = prostate.hex, family = "gaussian", nfolds = 5, alpha = 0.1)
K-Means
K means is a clustering algorithm that allows users to characterize data. This algorithm does not rely on a dependent variable. For information on the K-Means algorithm see K-Means
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath)
prostate.km = h2o.kmeans(data = prostate.hex, centers = 10, cols = c("AGE", "RACE", "VOL", "GLEASON"))
print(prostate.km)
Principal Components Analysis
Principal Components Analysis maps a set of variables onto a subspace via linear transformations. PCA is the first step in Principal Components Regression. For more information on PCA see Principal Components Analysis.
ausPath = system.file("extdata", "australia.csv", package="h2o")
australia.hex = h2o.importFile(localH2O, path = ausPath)
australia.pca = h2o.prcomp(data = australia.hex, standardize = TRUE)
print(australia.pca)
summary(australia.pca)
Principal Components Regression
PCR is an algorithm that allows users to map a set of variables to a new set of linearly independent variables. The new set of variables are linearly independent linear combinations of the original variables and exist in a subspace of lower dimension. This transformation is then prepended to a regression model, often improving results. For more information on PCA see Principal Components Analysis.
prostate.hex = h2o.importFile(localH2O, path =
"https://raw.github.com/0xdata/h2o/master/smalldata/logreg/prostate.csv",
key = "prostate.hex")
h2o.pcr(x = c("AGE","RACE","PSA","DCAPS"), y = "CAPSULE", data =
prostate.hex, family = "binomial",
nfolds = 10, alpha = 0.5, ncomp = 3)
Obtaining Predictions¶
Predict
Used to apply an H2O model to a holdout set to obtain predictions based on model results. In the examples below models are first generated, and then the predictions for that model are obtained.
prostate.hex = h2o.importFile(localH2O, path =
"https://raw.github.com/0xdata/h2o/master/smalldata/logreg/prostate.csv", key = "prostate.hex")
prostate.glm = h2o.glm(y = "CAPSULE", x =
c("AGE","RACE","PSA","DCAPS"), data = prostate.hex,
family = "binomial", nfolds = 10, alpha = 0.5)
prostate.fit = h2o.predict(object = prostate.glm, newdata = prostate.hex)
summary(prostate.fit)
Other Useful Functions¶
Get Frame
For users that alternate between using the web interface and the R API or for multiple users accessing the same H2O, this function gives the user the option to create a reference object for a data frame sitting in H2O. Assuming there’s a prostate.hex sitting in the KV store.
prostate.hex = h2o.getFrame(h2o = localH2O, key = "prostate.hex")
summary(prostate.hex)
Get Model
For users that alternate between using the web interface and the R API, this function gives the user the option create a reference object for a data frame sitting in H2O. Assuming there’s a GLMModel__ba724fe4f6d6d5b8b6370f776df94e47 model sitting in the KV store.
glm.model = h2o.getModel(h2o = localH2O, key = "GLMModel__ba724fe4f6d6d5b8b6370f776df94e47")
glm.model
List all H2O Objects
Used to generate a list of all H2O objects that have been generated during a work session, along with each objects byte size.
prostate.hex = h2o.importFile(localH2O, path = prosPath, key = "prostate.hex")
prostate.split = h2o.splitFrame(prostate.hex , ratio = 0.8)
prostate.train = prostate.split[1]
prostate.train = h2o.assign(prostate.train, "prostate.train")
h2o.ls(localH2O)
Remove an H2O object from the server where H2O is running
Users may wish to remove an H2O object on the server that is associated with an object in the R environment. Recommended behavior is to also remove the object in the R environment.
localH2O = h2o.init()
prosPath = system.file("extdata", "prostate.csv", package="h2o")
prostate.hex = h2o.importFile(localH2O, path = prosPath, key = "prostate.hex")
prostate.split = h2o.splitFrame(prostate.hex , ratio = 0.8)
prostate.train = prostate.split[1]
prostate.train = h2o.assign(prostate.train, "prostate.train")
h2o.ls(localH2O)
h2o.rm(object= localH2O, keys= "prostate.train")
h2o.ls(localH2O)